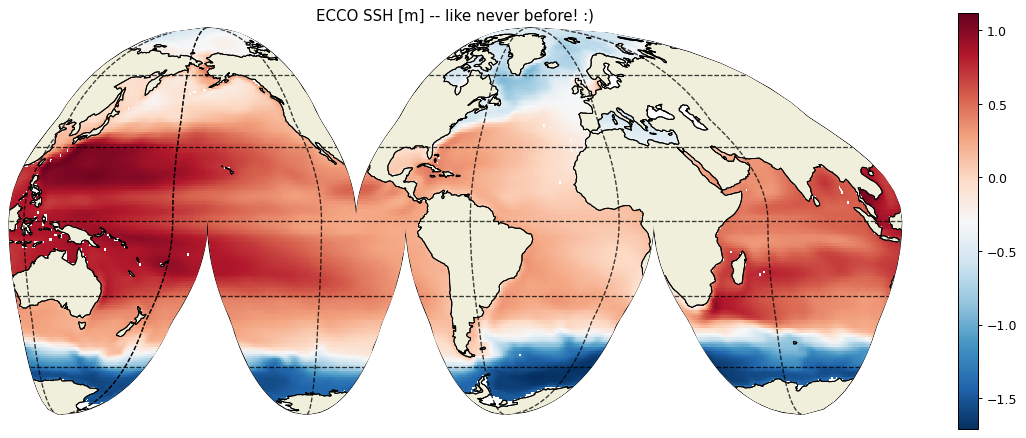
Plotting Tiles¶
Objectives¶
Introduce several different methods for plotting ECCO v4 fields that are stored as tiles in Datasets or DataArrays. Emphasis is placed on fields stored on the ECCO v4 native llc90 grid and loaded from NetCDF tile files.
Introduction¶
“Over the years many different plotting modules and packages have been developed for Python. For most of that time there was no clear favorite package, but recently matplotlib has become the most widely used. Nevertheless, many of the others are still available and may suit your tastes or needs better. Some of these are interfaces to existing plotting libraries while others are Python-centered new implementations. – from : https://wiki.python.org/moin/NumericAndScientific/Plotting
The link above profiles a long list of Python tools for plotting. In this tutorial we use just two libraries, matplotlib and Cartopy.
Note: In this tutorial you will need to have downloaded monthly SSH, THETA, and SALT for the year 2000. The ShortNames of the datasets needed are ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4 and ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4. You will also need the grid file.
matplotlib¶
“Matplotlib is a Python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments across platforms. Matplotlib can be used in Python scripts, the Python and IPython shell, the jupyter notebook, web application servers, and four graphical user interface toolkits.”
“For simple plotting the pyplot module provides a MATLAB-like interface, particularly when combined with [Juypter Notebooks]. For the power user, you have full control of line styles, font properties, axes properties, etc, via an object oriented interface or via a set of functions familiar to MATLAB users.” – from https://matplotlib.org/index.html
Matplotlib and pyplot even have a tutorial: https://matplotlib.org/users/pyplot_tutorial.html
Cartopy¶
“Cartopy is a Python package designed for geospatial data processing in order to produce maps and other geospatial data analyses.”
Cartopy makes use of the powerful PROJ.4, NumPy and Shapely libraries and includes a programmatic interface built on top of Matplotlib for the creation of publication quality maps.
Key features of cartopy are its object oriented projection definitions, and its ability to transform points, lines, vectors, polygons and images between those projections.
You will find cartopy especially useful for large area / small scale data, where Cartesian assumptions of spherical data traditionally break down. If you’ve ever experienced a singularity at the pole or a cut-off at the dateline, it is likely you will appreciate cartopy’s unique features!”*
The default orientation of the lat-lon-cap tile fields¶
Before we begin plotting ECCOv4 fields on the native llc90 model grid we are reminded how how the 13 tiles are oriented with respect to their “local” x and y and with respect to each other.
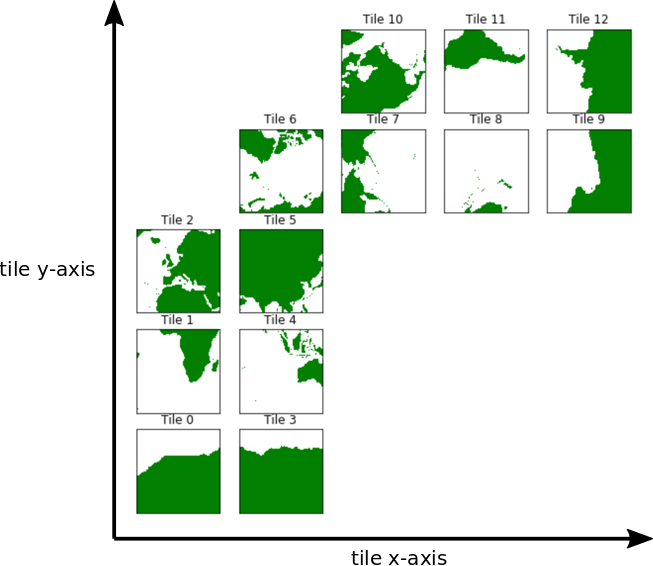
Tiles 7-12 are rotated 90 degrees counter-clockwise relative to tiles 0-5.
Note: The rotated orientation of tiles 7-12 presents some complications but don’t panic! The good news is that you don’t need to reorient tiles to plot them.
Plotting single tiles using imshow, pcolormesh, and contourf¶
First, let’s load the all 13 tiles for sea surface height and the model grid parameters.
[1]:
import numpy as np
import sys
import xarray as xr
import matplotlib.pyplot as plt
%matplotlib inline
import glob
import warnings
warnings.filterwarnings('ignore')
[2]:
# load some useful cartopy routines
from cartopy import config
import cartopy.crs as ccrs
import cartopy.feature as cfeature
# and a new matplotlib routine
import matplotlib.path as mpath
[3]:
## Import the ecco_v4_py library into Python
## =========================================
## If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. The example below adds
## ecco_v4_py to the user's path if it is stored in the folder
## ECCOv4-py under the user's home directory
from os.path import join,expanduser
user_home_dir = expanduser('~')
sys.path.append(join(user_home_dir,'ECCOv4-py'))
import ecco_v4_py as ecco
[4]:
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to ~/Downloads/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join(user_home_dir,'Downloads','ECCO_V4r4_PODAAC')
[5]:
## Load the model grid
ecco_grid = xr.open_dataset(glob.glob(join(ECCO_dir,'*GEOMETRY*','*.nc'))[0])
## Load one year of 2D monthly data: SSH, temperature and salinity
ds_SSH = xr.open_mfdataset(join(ECCO_dir,'*SSH*MONTHLY*','*_2000-*.nc'))
ds_temp_sal = xr.open_mfdataset(join(ECCO_dir,'*TEMP*SAL*MONTHLY*','*_2000-*.nc'))
## select only *surface* temperature and salinity (SST and SSS)
ds_SST_SSS = ds_temp_sal.isel(k=0)
## Copy ecco_ds from ecco_grid dataset
ecco_ds = ecco_grid.copy()
## Add SSH, SST, and SSS variables to ecco_ds
ecco_ds['SSH'] = ds_SSH['SSH']
ecco_ds['SST'] = ds_SST_SSS['THETA']
ecco_ds['SSS'] = ds_SST_SSS['SALT']
## Load ecco_ds into memory
ecco_ds = ecco_ds.load()
[6]:
ecco_ds
[6]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50, k_p1: 51, tile: 13, nb: 4, nv: 2, time: 12)
Coordinates: (12/21)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 -10.0 -20.0 -30.0 ... -5.678e+03 -6.134e+03
Zl (k_l) float32 0.0 -10.0 -20.0 ... -4.834e+03 -5.244e+03 -5.678e+03
XC_bnds (tile, j, i, nb) float32 -115.0 -115.0 -107.9 ... -115.0 -108.5
YC_bnds (tile, j, i, nb) float32 -88.18 -88.32 -88.3 ... -88.18 -88.16
Z_bnds (k, nv) float32 0.0 -10.0 -10.0 ... -5.678e+03 -6.134e+03
* time (time) datetime64[ns] 2000-01-16T12:00:00 ... 2000-12-16T12:00:00
Dimensions without coordinates: nb, nv
Data variables: (12/24)
CS (tile, j, i) float32 0.06158 0.06675 0.07293 ... -0.9854 -0.9984
SN (tile, j, i) float32 -0.9981 -0.9978 -0.9973 ... -0.1705 -0.05718
rA (tile, j, i) float32 3.623e+08 3.633e+08 ... 3.685e+08 3.611e+08
dxG (tile, j_g, i) float32 1.558e+04 1.559e+04 ... 2.36e+04 2.314e+04
dyG (tile, j, i_g) float32 2.321e+04 2.327e+04 ... 1.56e+04 1.558e+04
Depth (tile, j, i) float32 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
... ...
maskC (k, tile, j, i) bool False False False False ... False False False
maskW (k, tile, j, i_g) bool False False False ... False False False
maskS (k, tile, j_g, i) bool False False False ... False False False
SSH (time, tile, j, i) float32 nan nan nan nan nan ... nan nan nan nan
SST (time, tile, j, i) float32 nan nan nan nan nan ... nan nan nan nan
SSS (time, tile, j, i) float32 nan nan nan nan nan ... nan nan nan nan
Attributes: (12/58)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
references: ECCO Consortium, Fukumori, I., Wang, O.,...
source: The ECCO V4r4 state estimate was produce...
standard_name_vocabulary: NetCDF Climate and Forecast (CF) Metadat...
summary: This dataset provides geometric paramete...
title: ECCO Geometry Parameters for the Lat-Lon...
uuid: 87ff7d24-86e5-11eb-9c5f-f8f21e2ee3e0- i: 90
- i_g: 90
- j: 90
- j_g: 90
- k: 50
- k_u: 50
- k_l: 50
- k_p1: 51
- tile: 13
- nb: 4
- nv: 2
- time: 12
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_u(k_u)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z corresponding to the bottom face of tracer grid cells ('w' locations)
- c_grid_axis_shift :
- 0.5
- swap_dim :
- Zu
- comment :
- First index corresponds to the bottom face of the uppermost tracer grid cell. The use of 'u' in the variable name follows the MITgcm convention for naming the bottom face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_l(k_l)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z corresponding to the top face of tracer grid cells ('w' locations)
- c_grid_axis_shift :
- -0.5
- swap_dim :
- Zl
- comment :
- First index corresponds to the top face of the uppermost tracer grid cell. The use of 'l' in the variable name follows the MITgcm convention for naming the top face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_p1(k_p1)int320 1 2 3 4 5 6 ... 45 46 47 48 49 50
- axis :
- Z
- long_name :
- grid index in z for variables at 'w' locations
- c_grid_axis_shift :
- [-0.5 0.5]
- swap_dim :
- Zp1
- comment :
- Includes top of uppermost model tracer cell (k_p1=0) and bottom of lowermost tracer cell (k_p1=51).
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- XC(tile, j, i)float32-111.6 -111.3 ... -105.6 -111.9
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-111.60647 , -111.303 , -110.94285 , ..., 64.791115, 64.80521 , 64.81917 ], [-104.8196 , -103.928444, -102.87706 , ..., 64.36745 , 64.41012 , 64.4524 ], [ -98.198784, -96.788055, -95.14185 , ..., 63.936497, 64.008224, 64.0793 ], ..., [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ]], [[ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], ... [ -40.5 , -40.5 , -40.5 , ..., -40.5 , -40.5 , -40.5 ], [ -39.5 , -39.5 , -39.5 , ..., -39.5 , -39.5 , -39.5 ], [ -38.5 , -38.5 , -38.5 , ..., -38.5 , -38.5 , -38.5 ]], [[-127.5 , -127.5 , -127.5 , ..., -115.850204, -115.50567 , -115.166985], [-126.5 , -126.5 , -126.5 , ..., -115.78025 , -115.464066, -115.153244], [-125.5 , -125.5 , -125.5 , ..., -115.71079 , -115.42275 , -115.139595], ..., [ -40.5 , -40.5 , -40.5 , ..., -101.42989 , -106.83081 , -112.28605 ], [ -39.5 , -39.5 , -39.5 , ..., -100.48844 , -106.24874 , -112.090065], [ -38.5 , -38.5 , -38.5 , ..., -99.42048 , -105.58465 , -111.86579 ]]], dtype=float32) - YC(tile, j, i)float32-88.24 -88.38 ... -88.08 -88.1
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.24259 , -88.382515 , -88.52242 , ..., -80.57383 , -80.50492 , -80.43992 ], [-88.21676 , -88.354485 , -88.49178 , ..., -80.56925 , -80.50038 , -80.43542 ], [-88.164764 , -88.29827 , -88.43065 , ..., -80.55984 , -80.49105 , -80.42617 ], ..., [-58.321964 , -58.321964 , -58.321964 , ..., -58.321964 , -58.321964 , -58.321964 ], [-57.79962 , -57.79962 , -57.79962 , ..., -57.79962 , -57.79962 , -57.79962 ], [-57.271408 , -57.271408 , -57.271408 , ..., -57.271408 , -57.271408 , -57.271408 ]], [[-56.73891 , -56.73891 , -56.73891 , ..., -56.73891 , -56.73891 , -56.73891 ], [-56.2021 , -56.2021 , -56.2021 , ..., -56.2021 , -56.2021 , -56.2021 ], [-55.65936 , -55.65936 , -55.65936 , ..., -55.65936 , -55.65936 , -55.65936 ], ... [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ]], [[-57.271408 , -57.79962 , -58.321964 , ..., -80.36532 , -80.3745 , -80.37896 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.30862 , -80.317726 , -80.32216 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.25618 , -80.26523 , -80.26963 ], ..., [-57.271408 , -57.79962 , -58.321964 , ..., -87.7596 , -87.80198 , -87.822876 ], [-57.271408 , -57.79962 , -58.321964 , ..., -87.89526 , -87.94044 , -87.96276 ], [-57.271408 , -57.79962 , -58.321964 , ..., -88.030365 , -88.07871 , -88.10267 ]]], dtype=float32) - XG(tile, j_g, i_g)float32-115.0 -115.0 ... -102.9 -109.0
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-115. , -115. , -115. , ..., 65. , 65. , 65. ], [-108.47213 , -107.915245, -107.26045 , ..., 64.56598 , 64.59443 , 64.62261 ], [-101.9954 , -100.91797 , -99.66073 , ..., 64.12406 , 64.18147 , 64.23835 ], ..., [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ]], [[ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], ... [ -41. , -41. , -41. , ..., -41. , -41. , -41. ], [ -40. , -40. , -40. , ..., -40. , -40. , -40. ], [ -39. , -39. , -39. , ..., -39. , -39. , -39. ]], [[-128. , -128. , -128. , ..., -116.06686 , -115.70526 , -115.34945 ], [-127. , -127. , -127. , ..., -115.982254, -115.64933 , -115.32173 ], [-126. , -126. , -126. , ..., -115.89828 , -115.59381 , -115.29422 ], ..., [ -41. , -41. , -41. , ..., -99.28857 , -104.466286, -109.73619 ], [ -40. , -40. , -40. , ..., -98.24634 , -103.74563 , -109.36924 ], [ -39. , -39. , -39. , ..., -97.07265 , -102.928925, -108.95171 ]]], dtype=float32) - YG(tile, j_g, i_g)float32-88.18 -88.32 ... -87.99 -88.02
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.17569 , -88.31587 , -88.45609 , ..., -80.610275, -80.53945 , -80.47247 ], [-88.16335 , -88.30251 , -88.44153 , ..., -80.608 , -80.5372 , -80.470245], [-88.12586 , -88.26202 , -88.397514, ..., -80.601006, -80.530266, -80.46337 ], ..., [-58.58002 , -58.58002 , -58.58002 , ..., -58.58002 , -58.58002 , -58.58002 ], [-58.06183 , -58.06183 , -58.06183 , ..., -58.06183 , -58.06183 , -58.06183 ], [-57.53605 , -57.53605 , -57.53605 , ..., -57.53605 , -57.53605 , -57.53605 ]], [[-57.005695, -57.005695, -57.005695, ..., -57.005695, -57.005695, -57.005695], [-56.471046, -56.471046, -56.471046, ..., -56.471046, -56.471046, -56.471046], [-55.931786, -55.931786, -55.931786, ..., -55.931786, -55.931786, -55.931786], ... [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046]], [[-57.005695, -57.53605 , -58.06183 , ..., -80.38878 , -80.400444, -80.40726 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.33003 , -80.34161 , -80.34837 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.27549 , -80.286995, -80.29371 ], ..., [-57.005695, -57.53605 , -58.06183 , ..., -87.66318 , -87.71477 , -87.7454 ], [-57.005695, -57.53605 , -58.06183 , ..., -87.797325, -87.85214 , -87.884766], [-57.005695, -57.53605 , -58.06183 , ..., -87.93075 , -87.9892 , -88.02409 ]]], dtype=float32) - Z(k)float32-5.0 -15.0 ... -5.906e+03
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([-5.000000e+00, -1.500000e+01, -2.500000e+01, -3.500000e+01, -4.500000e+01, -5.500000e+01, -6.500000e+01, -7.500500e+01, -8.502500e+01, -9.509500e+01, -1.053100e+02, -1.158700e+02, -1.271500e+02, -1.397400e+02, -1.544700e+02, -1.724000e+02, -1.947350e+02, -2.227100e+02, -2.574700e+02, -2.999300e+02, -3.506800e+02, -4.099300e+02, -4.774700e+02, -5.527100e+02, -6.347350e+02, -7.224000e+02, -8.144700e+02, -9.097400e+02, -1.007155e+03, -1.105905e+03, -1.205535e+03, -1.306205e+03, -1.409150e+03, -1.517095e+03, -1.634175e+03, -1.765135e+03, -1.914150e+03, -2.084035e+03, -2.276225e+03, -2.491250e+03, -2.729250e+03, -2.990250e+03, -3.274250e+03, -3.581250e+03, -3.911250e+03, -4.264250e+03, -4.640250e+03, -5.039250e+03, -5.461250e+03, -5.906250e+03], dtype=float32) - Zp1(k_p1)float320.0 -10.0 ... -5.678e+03 -6.134e+03
- long_name :
- depth of top/bottom face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- Contains one element more than the number of vertical layers. First element is 0m, the depth of the top face of the uppermost grid cell. Last element is the depth of the bottom face of the deepest grid cell.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zu(k_u)float32-10.0 -20.0 ... -6.134e+03
- long_name :
- depth of bottom face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- First element is -10m, the depth of the bottom face of the uppermost tracer grid cell. Last element is the depth of the bottom face of the deepest grid cell. The use of 'u' in the variable name follows the MITgcm convention for naming the bottom face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zl(k_l)float320.0 -10.0 ... -5.244e+03 -5.678e+03
- long_name :
- depth of top face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- First element is 0m, the depth of the top face of the uppermost tracer grid cell (i.e., the ocean surface). Last element is the depth of the top face of the deepest grid cell. The use of 'l' in the variable name follows the MITgcm convention for naming the top face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. ], dtype=float32) - XC_bnds(tile, j, i, nb)float32-115.0 -115.0 ... -115.0 -108.5
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
array([[[[-115. , -115. , -107.915245, -108.47213 ], [-115. , -115. , -107.26045 , -107.915245], [-115. , -115. , -106.47914 , -107.26045 ], ..., [ 65. , 65. , 64.59443 , 64.56598 ], [ 65. , 65. , 64.62261 , 64.59443 ], [ 65. , 65. , 64.65055 , 64.62261 ]], [[-108.47213 , -107.915245, -100.91797 , -101.9954 ], [-107.915245, -107.26045 , -99.66073 , -100.91797 ], [-107.26045 , -106.47914 , -98.17517 , -99.66073 ], ..., [ 64.56598 , 64.59443 , 64.18147 , 64.12406 ], [ 64.59443 , 64.62261 , 64.23835 , 64.18147 ], [ 64.62261 , 64.65055 , 64.29474 , 64.23835 ]], [[-101.9954 , -100.91797 , -94.218956, -95.74128 ], [-100.91797 , -99.66073 , -92.46296 , -94.218956], [ -99.66073 , -98.17517 , -90.417885, -92.46296 ], ..., ... [ -99.28857 , -104.466286, -103.74563 , -98.24634 ], [-104.466286, -109.73619 , -109.36924 , -103.74563 ], [-109.73619 , -115. , -115. , -109.36924 ]], [[ -40. , -40. , -39. , -39. ], [ -40. , -40. , -39. , -39. ], [ -40. , -40. , -39. , -39. ], ..., [ -98.24634 , -103.74563 , -102.928925, -97.07265 ], [-103.74563 , -109.36924 , -108.95171 , -102.928925], [-109.36924 , -115. , -115. , -108.95171 ]], [[ -39. , -39. , -38. , -38. ], [ -39. , -39. , -38. , -38. ], [ -39. , -39. , -38. , -38. ], ..., [ -97.07265 , -102.928925, -101.9954 , -95.74128 ], [-102.928925, -108.95171 , -108.47213 , -101.9954 ], [-108.95171 , -115. , -115. , -108.47213 ]]]], dtype=float32) - YC_bnds(tile, j, i, nb)float32-88.18 -88.32 ... -88.18 -88.16
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
array([[[[-88.17569 , -88.31587 , -88.30251 , -88.16335 ], [-88.31587 , -88.45609 , -88.44153 , -88.30251 ], [-88.45609 , -88.596344, -88.580345, -88.44153 ], ..., [-80.610275, -80.53945 , -80.5372 , -80.608 ], [-80.53945 , -80.47247 , -80.470245, -80.5372 ], [-80.47247 , -80.40948 , -80.40726 , -80.470245]], [[-88.16335 , -88.30251 , -88.26202 , -88.12586 ], [-88.30251 , -88.44153 , -88.397514, -88.26202 ], [-88.44153 , -88.580345, -88.53216 , -88.397514], ..., [-80.608 , -80.5372 , -80.530266, -80.601006], [-80.5372 , -80.470245, -80.46337 , -80.530266], [-80.470245, -80.40726 , -80.400444, -80.46337 ]], [[-88.12586 , -88.26202 , -88.194695, -88.06328 ], [-88.26202 , -88.397514, -88.32474 , -88.194695], [-88.397514, -88.53216 , -88.453026, -88.32474 ], ..., ... [-87.66318 , -87.71477 , -87.85214 , -87.797325], [-87.71477 , -87.7454 , -87.884766, -87.85214 ], [-87.7454 , -87.75544 , -87.89547 , -87.884766]], [[-57.005695, -57.53605 , -57.53605 , -57.005695], [-57.53605 , -58.06183 , -58.06183 , -57.53605 ], [-58.06183 , -58.58002 , -58.58002 , -58.06183 ], ..., [-87.797325, -87.85214 , -87.9892 , -87.93075 ], [-87.85214 , -87.884766, -88.02409 , -87.9892 ], [-87.884766, -87.89547 , -88.03555 , -88.02409 ]], [[-57.005695, -57.53605 , -57.53605 , -57.005695], [-57.53605 , -58.06183 , -58.06183 , -57.53605 ], [-58.06183 , -58.58002 , -58.58002 , -58.06183 ], ..., [-87.93075 , -87.9892 , -88.12586 , -88.06328 ], [-87.9892 , -88.02409 , -88.16335 , -88.12586 ], [-88.02409 , -88.03555 , -88.17569 , -88.16335 ]]]], dtype=float32) - Z_bnds(k, nv)float320.0 -10.0 ... -5.678e+03 -6.134e+03
- comment :
- One pair of depths for each vertical level.
- coverage_content_type :
- coordinate
- long_name :
- depths of top and bottom faces of tracer grid cell
array([[ 0. , -10. ], [ -10. , -20. ], [ -20. , -30. ], [ -30. , -40. ], [ -40. , -50. ], [ -50. , -60. ], [ -60. , -70. ], [ -70. , -80.01 ], [ -80.01 , -90.04 ], [ -90.04 , -100.15 ], [ -100.15 , -110.47 ], [ -110.47 , -121.270004], [ -121.270004, -133.03 ], [ -133.03 , -146.45 ], [ -146.45 , -162.48999 ], [ -162.48999 , -182.31 ], [ -182.31 , -207.16 ], [ -207.16 , -238.26001 ], [ -238.26001 , -276.68 ], [ -276.68 , -323.18 ], ... [-1155.53 , -1255.54 ], [-1255.54 , -1356.87 ], [-1356.87 , -1461.4299 ], [-1461.4299 , -1572.7599 ], [-1572.7599 , -1695.5898 ], [-1695.5898 , -1834.6798 ], [-1834.6798 , -1993.6199 ], [-1993.6199 , -2174.45 ], [-2174.45 , -2378. ], [-2378. , -2604.5 ], [-2604.5 , -2854. ], [-2854. , -3126.5 ], [-3126.5 , -3422. ], [-3422. , -3740.5 ], [-3740.5 , -4082. ], [-4082. , -4446.5 ], [-4446.5 , -4834. ], [-4834. , -5244.5 ], [-5244.5 , -5678. ], [-5678. , -6134.5 ]], dtype=float32) - time(time)datetime64[ns]2000-01-16T12:00:00 ... 2000-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2000-01-16T12:00:00.000000000', '2000-02-15T12:00:00.000000000', '2000-03-16T12:00:00.000000000', '2000-04-16T00:00:00.000000000', '2000-05-16T12:00:00.000000000', '2000-06-16T00:00:00.000000000', '2000-07-16T12:00:00.000000000', '2000-08-16T12:00:00.000000000', '2000-09-16T00:00:00.000000000', '2000-10-16T12:00:00.000000000', '2000-11-16T00:00:00.000000000', '2000-12-16T12:00:00.000000000'], dtype='datetime64[ns]')
- CS(tile, j, i)float320.06158 0.06675 ... -0.9854 -0.9984
- long_name :
- cosine of tracer grid cell orientation vs geographical north
- units :
- 1
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- comment :
- CS and SN are required to calculate the geographic (meridional, zonal) components of vectors on the curvilinear model grid. Note: for vector R with components R_x and R_y: R_{east} = CS R_x - SN R_y. R_{north} = SN R_x + CS R_y
array([[[ 6.15781285e-02, 6.67537600e-02, 7.29308128e-02, ..., 3.29420231e-02, 3.47638950e-02, 3.68907340e-02], [ 1.83196113e-01, 1.98062390e-01, 2.15639561e-01, ..., 9.90367457e-02, 1.04462028e-01, 1.10784896e-01], [ 2.99260259e-01, 3.21938545e-01, 3.48297477e-01, ..., 1.65254667e-01, 1.74131751e-01, 1.84442252e-01], ..., [ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00], [ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00], [ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00]], [[ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00], [ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00], [ 1.00000000e+00, 1.00000000e+00, 1.00000000e+00, ..., 1.00000000e+00, 1.00000000e+00, 1.00000000e+00], ... -6.34209863e-15, -6.34209863e-15, 6.52405994e-15], [ 8.99243529e-16, 1.79623861e-15, 1.34452388e-15, ..., -0.00000000e+00, -0.00000000e+00, -6.52405994e-15], [ 2.55257101e-18, -8.99243529e-16, -4.47528799e-16, ..., 1.25081762e-14, -0.00000000e+00, -0.00000000e+00]], [[-0.00000000e+00, 1.34317132e-14, 1.34317132e-14, ..., 9.80502069e-01, 9.92987871e-01, 9.99223709e-01], [ 6.52405994e-15, -1.34317132e-14, -6.61670417e-15, ..., 9.77538407e-01, 9.91897821e-01, 9.99101698e-01], [-6.52405994e-15, -0.00000000e+00, -6.81500898e-15, ..., 9.73717213e-01, 9.90482509e-01, 9.98942673e-01], ..., [ 6.52405994e-15, -0.00000000e+00, 6.81500898e-15, ..., -9.68940675e-01, -9.88701105e-01, -9.98742878e-01], [-6.52405994e-15, 1.34317132e-14, 6.61670417e-15, ..., -9.64956880e-01, -9.87200499e-01, -9.98572946e-01], [-0.00000000e+00, -1.34317132e-14, -1.34317132e-14, ..., -9.60118830e-01, -9.85361755e-01, -9.98363793e-01]]], dtype=float32) - SN(tile, j, i)float32-0.9981 -0.9978 ... -0.05718
- long_name :
- sine of tracer grid cell orientation vs geographical north
- units :
- 1
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- comment :
- CS and SN are required to calculate the geographic (meridional, zonal) components of vectors on the curvilinear model grid. Note: for vector R with components R_x and R_y in local grid directions x and y, the geographical eastward component R_{east} = CS R_x - SN R_y. The geographical northward component R_{north} = SN R_x + CS R_y.
array([[[-9.9810225e-01, -9.9776947e-01, -9.9733698e-01, ..., 9.9945724e-01, 9.9939555e-01, 9.9931931e-01], [-9.8307639e-01, -9.8018944e-01, -9.7647303e-01, ..., 9.9508375e-01, 9.9452889e-01, 9.9384439e-01], [-9.5417154e-01, -9.4676059e-01, -9.3738407e-01, ..., 9.8625094e-01, 9.8472238e-01, 9.8284334e-01], ..., [ 1.3431713e-14, -6.6167042e-15, -6.8150090e-15, ..., 6.8150090e-15, 6.6167042e-15, -1.3431713e-14], [ 1.3431713e-14, -1.3431713e-14, -0.0000000e+00, ..., -0.0000000e+00, 1.3431713e-14, -1.3431713e-14], [-0.0000000e+00, 6.5240599e-15, -6.5240599e-15, ..., 6.5240599e-15, -6.5240599e-15, -0.0000000e+00]], [[-0.0000000e+00, 6.5240599e-15, -6.5240599e-15, ..., 6.5240599e-15, -6.5240599e-15, -0.0000000e+00], [-0.0000000e+00, -0.0000000e+00, 6.3420986e-15, ..., -6.3420986e-15, -0.0000000e+00, -0.0000000e+00], [-1.2508176e-14, -0.0000000e+00, 6.3420986e-15, ..., -6.3420986e-15, -0.0000000e+00, 1.2508176e-14], ... [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -1.0000000e+00, -1.0000000e+00, -1.0000000e+00], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -1.0000000e+00, -1.0000000e+00, -1.0000000e+00], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -1.0000000e+00, -1.0000000e+00, -1.0000000e+00]], [[-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -1.9650881e-01, -1.1821613e-01, -3.9395642e-02], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -2.1075734e-01, -1.2703830e-01, -4.2377368e-02], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -2.2776045e-01, -1.3763885e-01, -4.5972619e-02], ..., [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -2.4729335e-01, -1.4990035e-01, -5.0126668e-02], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -2.6240838e-01, -1.5948406e-01, -5.3404655e-02], [-1.0000000e+00, -1.0000000e+00, -1.0000000e+00, ..., -2.7959219e-01, -1.7047660e-01, -5.7182025e-02]]], dtype=float32) - rA(tile, j, i)float323.623e+08 3.633e+08 ... 3.611e+08
- long_name :
- area of tracer grid cell
- units :
- m2
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
array([[[3.6225645e+08, 3.6330096e+08, 3.6425389e+08, ..., 6.0276308e+07, 5.3553676e+07, 4.7093872e+07], [3.6961037e+08, 3.7067478e+08, 3.7164586e+08, ..., 6.1906760e+07, 5.5057284e+07, 4.8475600e+07], [3.7716797e+08, 3.7825142e+08, 3.7923990e+08, ..., 6.4021868e+07, 5.7052416e+07, 5.0355492e+07], ..., [3.3636692e+09, 3.3636692e+09, 3.3636692e+09, ..., 3.3636692e+09, 3.3636692e+09, 3.3636692e+09], [3.4631982e+09, 3.4631982e+09, 3.4631982e+09, ..., 3.4631982e+09, 3.4631982e+09, 3.4631982e+09], [3.5442668e+09, 3.5442668e+09, 3.5442668e+09, ..., 3.5442668e+09, 3.5442668e+09, 3.5442668e+09]], [[3.6245120e+09, 3.6245120e+09, 3.6245120e+09, ..., 3.6245120e+09, 3.6245120e+09, 3.6245120e+09], [3.7078205e+09, 3.7078205e+09, 3.7078205e+09, ..., 3.7078205e+09, 3.7078205e+09, 3.7078205e+09], [3.8142915e+09, 3.8142915e+09, 3.8142915e+09, ..., 3.8142915e+09, 3.8142915e+09, 3.8142915e+09], ... [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09], [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09], [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09]], [[3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 4.3963860e+07, 4.2193932e+07, 4.0928504e+07], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7910156e+07, 3.6244328e+07, 3.5089036e+07], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.2226466e+07, 3.0658362e+07, 2.9606462e+07], ..., [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7333379e+08, 3.6584362e+08, 3.5856013e+08], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7471066e+08, 3.6719626e+08, 3.5988749e+08], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7598819e+08, 3.6845133e+08, 3.6111910e+08]]], dtype=float32) - dxG(tile, j_g, i)float321.558e+04 1.559e+04 ... 2.314e+04
- long_name :
- distance between 'southwest' and 'southeast' corners of the tracer grid cell
- units :
- m
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'south' side of tracer grid cell. Note: 'south', 'southwest', and 'southeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[ 15584.907 , 15589.316 , 15593.201 , ..., 7873.7295, 7446.052 , 7004.153 ], [ 15588.651 , 15593.048 , 15596.92 , ..., 7888.4585, 7461.8286, 7021.1333], [ 15600.185 , 15604.543 , 15608.381 , ..., 7933.642 , 7510.2056, 7073.169 ], ..., [ 57957.625 , 57957.625 , 57957.625 , ..., 57957.625 , 57957.625 , 57957.625 ], [ 58813.305 , 58813.305 , 58813.305 , ..., 58813.305 , 58813.305 , 58813.305 ], [ 59676.61 , 59676.61 , 59676.61 , ..., 59676.61 , 59676.61 , 59676.61 ]], [[ 60542.324 , 60542.324 , 60542.324 , ..., 60542.324 , 60542.324 , 60542.324 ], [ 61409.805 , 61409.805 , 61409.805 , ..., 61409.805 , 61409.805 , 61409.805 ], [ 62279.344 , 62279.344 , 62279.344 , ..., 62279.344 , 62279.344 , 62279.344 ], ... [108086.08 , 106469.66 , 103581.64 , ..., 60816.504 , 59953.41 , 59441.125 ], [108086.08 , 106469.66 , 103581.64 , ..., 60816.504 , 59953.41 , 59441.125 ], [108086.08 , 106469.66 , 103581.64 , ..., 60816.504 , 59953.41 , 59441.125 ]], [[ 58963.117 , 58455.164 , 57611.023 , ..., 6833.7446, 6639.343 , 6479.438 ], [ 58963.117 , 58455.164 , 57611.023 , ..., 6347.175 , 6156.099 , 6002.6626], [ 58963.117 , 58455.164 , 57611.023 , ..., 5857.2334, 5668.565 , 5521.185 ], ..., [ 58963.117 , 58455.164 , 57611.023 , ..., 23901.465 , 23445.639 , 22990.21 ], [ 58963.117 , 58455.164 , 57611.023 , ..., 23982.959 , 23525.693 , 23068.766 ], [ 58963.117 , 58455.164 , 57611.023 , ..., 24059.047 , 23600.436 , 23142.107 ]]], dtype=float32) - dyG(tile, j, i_g)float322.321e+04 2.327e+04 ... 1.558e+04
- long_name :
- distance between 'southwest' and 'northwest' corners of the tracer grid cell
- units :
- m
- coordinate :
- YC XG
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'west' side of tracer grid cell. Note: 'west, 'southwest', and 'northwest' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[ 23210.262 , 23273.26 , 23331.125 , ..., 7878.489 , 7417.633 , 6951.1924], [ 23669.895 , 23734.096 , 23793.066 , ..., 8059.287 , 7591.293 , 7117.8667], [ 24129.756 , 24195.113 , 24255.148 , ..., 8267.303 , 7794.305 , 7316.3003], ..., [ 57611.023 , 57611.023 , 57611.023 , ..., 57611.023 , 57611.023 , 57611.023 ], [ 58455.164 , 58455.164 , 58455.164 , ..., 58455.164 , 58455.164 , 58455.164 ], [ 58963.117 , 58963.117 , 58963.117 , ..., 58963.117 , 58963.117 , 58963.117 ]], [[ 59441.125 , 59441.125 , 59441.125 , ..., 59441.125 , 59441.125 , 59441.125 ], [ 59953.41 , 59953.41 , 59953.41 , ..., 59953.41 , 59953.41 , 59953.41 ], [ 60816.504 , 60816.504 , 60816.504 , ..., 60816.504 , 60816.504 , 60816.504 ], ... [109498.625 , 109809.445 , 110084.7 , ..., 63155.766 , 62279.344 , 61409.805 ], [109498.625 , 109809.445 , 110084.7 , ..., 63155.766 , 62279.344 , 61409.805 ], [109498.625 , 109809.445 , 110084.7 , ..., 63155.766 , 62279.344 , 61409.805 ]], [[ 60542.324 , 59676.61 , 58813.305 , ..., 6718.4214, 6623.084 , 6566.8154], [ 60542.324 , 59676.61 , 58813.305 , ..., 6264.1724, 6160.646 , 6099.426 ], [ 60542.324 , 59676.61 , 58813.305 , ..., 5799.8853, 5686.719 , 5619.632 ], ..., [ 60542.324 , 59676.61 , 58813.305 , ..., 15603.497 , 15583.506 , 15571.836 ], [ 60542.324 , 59676.61 , 58813.305 , ..., 15609.626 , 15589.719 , 15578.099 ], [ 60542.324 , 59676.61 , 58813.305 , ..., 15615.09 , 15595.26 , 15583.685 ]]], dtype=float32) - Depth(tile, j, i)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- model seafloor depth below ocean surface at rest
- units :
- m
- coordinate :
- XC YC
- coverage_content_type :
- modelResult
- standard_name :
- sea_floor_depth_below_geoid
- comment :
- Model sea surface height (SSH) of 0m corresponds to an ocean surface at rest relative to the geoid. Depth corresponds to seafloor depth below geoid. Note: the MITgcm used by ECCO V4r4 implements 'partial cells' so the actual model seafloor depth may differ from the seafloor depth provided by the input bathymetry file.
array([[[ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [2977.8345 , 2503.9146 , 2562.9644 , ..., 5431.1567 , 5356.7544 , 5331.2 ], [3125.5176 , 2908.5 , 3046.7676 , ..., 5440.6733 , 5383.5386 , 5332.443 ], [3203.0798 , 3283.116 , 3312.7979 , ..., 5455.109 , 5433.769 , 5367.7656 ]], [[3284.1084 , 3485.7 , 3485.7 , ..., 5455.109 , 5444.462 , 5387.942 ], [3350.4321 , 3403.6208 , 3485.7 , ..., 5367.847 , 5379.0474 , 5359.3574 ], [3324.7493 , 3126.5 , 2243.637 , ..., 5244.5 , 5244.5 , 5196.211 ], ... [3384.1433 , 3728.414 , 4524. , ..., 3185.6 , 3107.3855 , 3246.3552 ], [3638.9377 , 3570.711 , 4352.1484 , ..., 3031.5972 , 2908.5 , 2908.5 ], [3965.616 , 3676.4983 , 4082. , ..., 3285.413 , 3066.3777 , 3055.8303 ]], [[3920.2725 , 3809.5889 , 3877.8499 , ..., 0. , 0. , 0. ], [3984.9038 , 3906.704 , 3950.57 , ..., 0. , 0. , 0. ], [3982.3193 , 3961.0894 , 4053.4758 , ..., 0. , 0. , 0. ], ..., [3374.1394 , 3362.2715 , 3243.364 , ..., 0. , 0. , 0. ], [2949.6958 , 3126.5 , 3213.2534 , ..., 0. , 0. , 0. ], [3076.3447 , 3119.1152 , 3126.5 , ..., 0. , 0. , 0. ]]], dtype=float32) - rAz(tile, j_g, i_g)float321.799e+08 1.805e+08 ... 3.642e+08
- long_name :
- area of vorticity 'g' grid cell
- units :
- m2
- coordinate :
- YG XG
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Vorticity cells are staggered in space relative to tracer cells, nominally situated on tracer cell corners. Vorticity cell (i,j) is located at the 'southwest' corner of tracer grid cell (i, j). Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[1.79944256e+08, 1.80486992e+08, 1.80983840e+08, ..., 3.16794380e+07, 2.82748640e+07, 2.49932220e+07], [3.65346336e+08, 3.66447584e+08, 3.67455744e+08, ..., 6.45169800e+07, 5.76093880e+07, 5.09512080e+07], [3.72789824e+08, 3.73911424e+08, 3.74938240e+08, ..., 6.64470160e+07, 5.94136880e+07, 5.26343600e+07], ..., [3.31241549e+09, 3.31241549e+09, 3.31241549e+09, ..., 3.31241549e+09, 3.31241549e+09, 3.31241549e+09], [3.41554739e+09, 3.41554739e+09, 3.41554739e+09, ..., 3.41554739e+09, 3.41554739e+09, 3.41554739e+09], [3.50457702e+09, 3.50457702e+09, 3.50457702e+09, ..., 3.50457702e+09, 3.50457702e+09, 3.50457702e+09]], [[3.58424499e+09, 3.58424499e+09, 3.58424499e+09, ..., 3.58424499e+09, 3.58424499e+09, 3.58424499e+09], [3.66506675e+09, 3.66506675e+09, 3.66506675e+09, ..., 3.66506675e+09, 3.66506675e+09, 3.66506675e+09], [3.75804186e+09, 3.75804186e+09, 3.75804186e+09, ..., 3.75804186e+09, 3.75804186e+09, 3.75804186e+09], ... [1.18844058e+10, 1.17948221e+10, 1.15813806e+10, ..., 3.87148928e+09, 3.75804186e+09, 3.66506675e+09], [1.18844058e+10, 1.17948221e+10, 1.15813806e+10, ..., 3.87148928e+09, 3.75804186e+09, 3.66506675e+09], [1.18844058e+10, 1.17948221e+10, 1.15813806e+10, ..., 3.87148928e+09, 3.75804186e+09, 3.66506675e+09]], [[3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 4.82358040e+07, 4.61414640e+07, 4.45743320e+07], [3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 4.19580600e+07, 3.99674760e+07, 3.85106320e+07], [3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 3.60372920e+07, 3.41445240e+07, 3.27916800e+07], ..., [3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 3.76430432e+08, 3.68841824e+08, 3.61469984e+08], [3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 3.77869632e+08, 3.70256544e+08, 3.62859008e+08], [3.58424499e+09, 3.50457702e+09, 3.41554739e+09, ..., 3.79207840e+08, 3.71572000e+08, 3.64150624e+08]]], dtype=float32) - dxC(tile, j, i_g)float321.558e+04 1.559e+04 ... 2.341e+04
- long_name :
- distance between centers of adjacent tracer grid cells in the 'x' direction
- units :
- m
- coordinate :
- YC XG
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'north' side of vorticity grid cells. Note: 'north' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[ 15583.418 , 15588.104 , 15592.241 , ..., 8085.6636, 7665.4624, 7230.907 ], [ 15591.017 , 15595.676 , 15599.79 , ..., 8114.4873, 7696.267 , 7263.974 ], [ 15606.636 , 15611.241 , 15615.306 , ..., 8173.435 , 7759.222 , 7331.496 ], ..., [ 58384.344 , 58384.344 , 58384.344 , ..., 58384.344 , 58384.344 , 58384.344 ], [ 59244.46 , 59244.46 , 59244.46 , ..., 59244.46 , 59244.46 , 59244.46 ], [ 60109.234 , 60109.234 , 60109.234 , ..., 60109.234 , 60109.234 , 60109.234 ]], [[ 60975.85 , 60975.85 , 60975.85 , ..., 60975.85 , 60975.85 , 60975.85 ], [ 61844.16 , 61844.16 , 61844.16 , ..., 61844.16 , 61844.16 , 61844.16 ], [ 62716.535 , 62716.535 , 62716.535 , ..., 62716.535 , 62716.535 , 62716.535 ], ... [108536.33 , 107413.53 , 105206.3 , ..., 61299.195 , 60340.285 , 59681.477 ], [108536.33 , 107413.53 , 105206.3 , ..., 61299.195 , 60340.285 , 59681.477 ], [108536.33 , 107413.53 , 105206.3 , ..., 61299.195 , 60340.285 , 59681.477 ]], [[ 59201.66 , 58725.49 , 58072.91 , ..., 6701.523 , 6489.9375, 6315.64 ], [ 59201.66 , 58725.49 , 58072.91 , ..., 6213.01 , 6002.6147, 5832.9194], [ 59201.66 , 58725.49 , 58072.91 , ..., 5722.2153, 5511.698 , 5345.8335], ..., [ 59201.66 , 58725.49 , 58072.91 , ..., 24171.275 , 23714.576 , 23258.152 ], [ 59201.66 , 58725.49 , 58072.91 , ..., 24250.736 , 23792.676 , 23334.83 ], [ 59201.66 , 58725.49 , 58072.91 , ..., 24324.758 , 23865.428 , 23406.256 ]]], dtype=float32) - dyC(tile, j_g, i)float321.156e+04 1.159e+04 ... 1.558e+04
- long_name :
- distance between centers of adjacent tracer grid cells in the 'y' direction
- units :
- m
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'east' side of vorticity grid cells. Note: 'east' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[ 11563.718 , 11593.785 , 11621.304 , ..., 3803.9153, 3573.0938, 3339.542 ], [ 23472.46 , 23533.473 , 23589.314 , ..., 7733.991 , 7266.5024, 6793.6157], [ 23932.863 , 23995.008 , 24051.885 , ..., 7925.047 , 7451.6045, 6973.0957], ..., [ 57150.88 , 57150.88 , 57150.88 , ..., 57150.88 , 57150.88 , 57150.88 ], [ 58072.91 , 58072.91 , 58072.91 , ..., 58072.91 , 58072.91 , 58072.91 ], [ 58725.49 , 58725.49 , 58725.49 , ..., 58725.49 , 58725.49 , 58725.49 ]], [[ 59201.66 , 59201.66 , 59201.66 , ..., 59201.66 , 59201.66 , 59201.66 ], [ 59681.477 , 59681.477 , 59681.477 , ..., 59681.477 , 59681.477 , 59681.477 ], [ 60340.285 , 60340.285 , 60340.285 , ..., 60340.285 , 60340.285 , 60340.285 ], ... [109658.375 , 109951.62 , 110208.53 , ..., 62716.535 , 61844.16 , 60975.85 ], [109658.375 , 109951.62 , 110208.53 , ..., 62716.535 , 61844.16 , 60975.85 ], [109658.375 , 109951.62 , 110208.53 , ..., 62716.535 , 61844.16 , 60975.85 ]], [[ 60109.234 , 59244.46 , 58384.344 , ..., 6890.8105, 6818.0396, 6782.365 ], [ 60109.234 , 59244.46 , 58384.344 , ..., 6437.876 , 6359.012 , 6320.301 ], [ 60109.234 , 59244.46 , 58384.344 , ..., 5973.588 , 5887.572 , 5845.279 ], ..., [ 60109.234 , 59244.46 , 58384.344 , ..., 15589.076 , 15573.266 , 15565.574 ], [ 60109.234 , 59244.46 , 58384.344 , ..., 15595.608 , 15579.867 , 15572.209 ], [ 60109.234 , 59244.46 , 58384.344 , ..., 15601.442 , 15585.765 , 15578.138 ]]], dtype=float32) - rAw(tile, j, i_g)float323.617e+08 3.628e+08 ... 3.648e+08
- long_name :
- area of 'v' grid cell
- units :
- m2
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Model 'v' grid cells are staggered in space between adjacent tracer grid cells in the 'x' direction. 'v' grid cell (i,j) is situated at the 'west' edge of tracer grid cell (i, j). Note: 'west' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[3.6169946e+08, 3.6279024e+08, 3.6378880e+08, ..., 6.3726332e+07, 5.6884112e+07, 5.0288944e+07], [3.6904275e+08, 3.7015430e+08, 3.7117190e+08, ..., 6.5421896e+07, 5.8450560e+07, 5.1730956e+07], [3.7659021e+08, 3.7772166e+08, 3.7875747e+08, ..., 6.7598592e+07, 6.0505120e+07, 5.3667840e+07], ..., [3.3636692e+09, 3.3636692e+09, 3.3636692e+09, ..., 3.3636692e+09, 3.3636692e+09, 3.3636692e+09], [3.4631982e+09, 3.4631982e+09, 3.4631982e+09, ..., 3.4631982e+09, 3.4631982e+09, 3.4631982e+09], [3.5442668e+09, 3.5442668e+09, 3.5442668e+09, ..., 3.5442668e+09, 3.5442668e+09, 3.5442668e+09]], [[3.6245120e+09, 3.6245120e+09, 3.6245120e+09, ..., 3.6245120e+09, 3.6245120e+09, 3.6245120e+09], [3.7078205e+09, 3.7078205e+09, 3.7078205e+09, ..., 3.7078205e+09, 3.7078205e+09, 3.7078205e+09], [3.8142915e+09, 3.8142915e+09, 3.8142915e+09, ..., 3.8142915e+09, 3.8142915e+09, 3.8142915e+09], ... [1.1884406e+10, 1.1794822e+10, 1.1581381e+10, ..., 3.8714893e+09, 3.7580419e+09, 3.6650668e+09], [1.1884406e+10, 1.1794822e+10, 1.1581381e+10, ..., 3.8714893e+09, 3.7580419e+09, 3.6650668e+09], [1.1884406e+10, 1.1794822e+10, 1.1581381e+10, ..., 3.8714893e+09, 3.7580419e+09, 3.6650668e+09]], [[3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 4.5054340e+07, 4.3012584e+07, 4.1501352e+07], [3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 3.8951032e+07, 3.7010132e+07, 3.5606112e+07], [3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 3.3220808e+07, 3.1374556e+07, 3.0071176e+07], ..., [3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 3.7716275e+08, 3.6956170e+08, 3.6217677e+08], [3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 3.7855126e+08, 3.7092659e+08, 3.6351690e+08], [3.5842450e+09, 3.5045770e+09, 3.4155474e+09, ..., 3.7983962e+08, 3.7219302e+08, 3.6476035e+08]]], dtype=float32) - rAs(tile, j_g, i)float321.802e+08 1.807e+08 ... 3.605e+08
- long_name :
- area of 'u' grid cell
- units :
- m2
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Model 'u' grid cells are staggered in space between adjacent tracer grid cells in the 'y' direction. 'u' grid cell (i,j) is situated at the 'south' edge of tracer grid cell (i, j). Note: 'south' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[1.8022141e+08, 1.8074112e+08, 1.8121526e+08, ..., 2.9962764e+07, 2.6617694e+07, 2.3403408e+07], [3.6590867e+08, 3.6696323e+08, 3.6792531e+08, ..., 6.1033988e+07, 5.4247132e+07, 4.7725612e+07], [3.7336256e+08, 3.7443661e+08, 3.7541648e+08, ..., 6.2900624e+07, 5.5990252e+07, 4.9350072e+07], ..., [3.3124155e+09, 3.3124155e+09, 3.3124155e+09, ..., 3.3124155e+09, 3.3124155e+09, 3.3124155e+09], [3.4155474e+09, 3.4155474e+09, 3.4155474e+09, ..., 3.4155474e+09, 3.4155474e+09, 3.4155474e+09], [3.5045770e+09, 3.5045770e+09, 3.5045770e+09, ..., 3.5045770e+09, 3.5045770e+09, 3.5045770e+09]], [[3.5842450e+09, 3.5842450e+09, 3.5842450e+09, ..., 3.5842450e+09, 3.5842450e+09, 3.5842450e+09], [3.6650668e+09, 3.6650668e+09, 3.6650668e+09, ..., 3.6650668e+09, 3.6650668e+09, 3.6650668e+09], [3.7580419e+09, 3.7580419e+09, 3.7580419e+09, ..., 3.7580419e+09, 3.7580419e+09, 3.7580419e+09], ... [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09], [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09], [1.1852359e+10, 1.1706299e+10, 1.1415347e+10, ..., 3.8142915e+09, 3.7078205e+09, 3.6245120e+09]], [[3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 4.7119472e+07, 4.5295252e+07, 4.3972408e+07], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 4.0892744e+07, 3.9175636e+07, 3.7966080e+07], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.5020080e+07, 3.3403948e+07, 3.2301228e+07], ..., [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7260762e+08, 3.6513021e+08, 3.5786006e+08], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7403475e+08, 3.6653222e+08, 3.5923587e+08], [3.5442668e+09, 3.4631982e+09, 3.3636692e+09, ..., 3.7536173e+08, 3.6783590e+08, 3.6051517e+08]]], dtype=float32) - drC(k_p1)float325.0 10.0 10.0 ... 422.0 445.0 228.2
- long_name :
- distance between the centers of adjacent tracer grid cells in the 'z' direction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- The first element corresponds to the distance between the depth of the center of the uppermost model grid cell and the surface.
array([ 5. , 10. , 10. , 10. , 10. , 10. , 10. , 10.005, 10.02 , 10.07 , 10.215, 10.56 , 11.28 , 12.59 , 14.73 , 17.93 , 22.335, 27.975, 34.76 , 42.46 , 50.75 , 59.25 , 67.54 , 75.24 , 82.025, 87.665, 92.07 , 95.27 , 97.415, 98.75 , 99.63 , 100.67 , 102.945, 107.945, 117.08 , 130.96 , 149.015, 169.885, 192.19 , 215.025, 238. , 261. , 284. , 307. , 330. , 353. , 376. , 399. , 422. , 445. , 228.25 ], dtype=float32) - drF(k)float3210.0 10.0 10.0 ... 433.5 456.5
- long_name :
- distance between the upper and lower interfaces of the model grid cell
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- cell_thickness
- comment :
- Nominal grid cell thickness. Note: in the z* coordinate system used in ECCO V4, actual tracer grid cell thickness, h, varies through time as h(i,j,k,t)= drF(k) hfacC(i,j,k,t).
array([ 10. , 10. , 10. , 10. , 10. , 10. , 10. , 10.01, 10.03, 10.11, 10.32, 10.8 , 11.76, 13.42, 16.04, 19.82, 24.85, 31.1 , 38.42, 46.5 , 55. , 63.5 , 71.58, 78.9 , 85.15, 90.18, 93.96, 96.58, 98.25, 99.25, 100.01, 101.33, 104.56, 111.33, 122.83, 139.09, 158.94, 180.83, 203.55, 226.5 , 249.5 , 272.5 , 295.5 , 318.5 , 341.5 , 364.5 , 387.5 , 410.5 , 433.5 , 456.5 ], dtype=float32) - PHrefC(k)float3249.05 147.1 ... 5.357e+04 5.794e+04
- long_name :
- reference ocean hydrostatic pressure at tracer grid cell center
- units :
- m2 s-2
- coverage_content_type :
- modelResult
- comment :
- PHrefC = p_ref (k) / rhoConst = rhoConst g z(k) / rhoConst = g z(k), where p_ref(k) is reference hydrostatic ocean pressure at center of tracer grid cell k, rhoConst is reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and z(k) is depth at center of tracer grid cell k. Units: p:[kg m-1 s-2], rhoConst:[kg m-3], g:[m s-2], z_m(t):[m]. Note: does not include atmospheric pressure loading. Quantity referred to in some contexts as hydrostatic pressure potential. PHIHYDcR is anomaly of PHrefC.
array([4.9049999e+01, 1.4714999e+02, 2.4525000e+02, 3.4335001e+02, 4.4145001e+02, 5.3954999e+02, 6.3765002e+02, 7.3579907e+02, 8.3409528e+02, 9.3288196e+02, 1.0330911e+03, 1.1366847e+03, 1.2473416e+03, 1.3708494e+03, 1.5153507e+03, 1.6912440e+03, 1.9103503e+03, 2.1847852e+03, 2.5257808e+03, 2.9423132e+03, 3.4401709e+03, 4.0214133e+03, 4.6839805e+03, 5.4220850e+03, 6.2267505e+03, 7.0867441e+03, 7.9899507e+03, 8.9245498e+03, 9.8801904e+03, 1.0848928e+04, 1.1826299e+04, 1.2813871e+04, 1.3823762e+04, 1.4882702e+04, 1.6031257e+04, 1.7315975e+04, 1.8777811e+04, 2.0444383e+04, 2.2329768e+04, 2.4439162e+04, 2.6773943e+04, 2.9334352e+04, 3.2120393e+04, 3.5132062e+04, 3.8369363e+04, 4.1832293e+04, 4.5520852e+04, 4.9435043e+04, 5.3574863e+04, 5.7940312e+04], dtype=float32) - PHrefF(k_p1)float320.0 98.1 ... 5.57e+04 6.018e+04
- long_name :
- reference ocean hydrostatic pressure at tracer grid cell top/bottom interface
- units :
- m2 s-2
- coverage_content_type :
- modelResult
- comment :
- PHrefF = p_ref (k_l) / rhoConst = rhoConst g z(k_l) / rhoConst = g z(k_l), where p_ref(k_l) is reference hydrostatic ocean pressure at lower interface of tracer grid cell k, rhoConst is reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and z(k) is depth at center of tracer grid cell k. Units: p:[kg m-1 s-2], rhoConst:[kg m-3], g:[m s-2], z_m(t):[m]. Note: does not include atmospheric pressure loading. Quantity referred to in some contexts as hydrostatic pressure potential. See PHrefC
array([ 0. , 98.1 , 196.2 , 294.3 , 392.4 , 490.5 , 588.6 , 686.7 , 784.8981, 883.2924, 982.4715, 1083.7107, 1189.6587, 1305.0243, 1436.6746, 1594.0269, 1788.461 , 2032.2396, 2337.3306, 2714.2307, 3170.3958, 3709.9458, 4332.881 , 5035.0806, 5809.09 , 6644.411 , 7529.0767, 8450.824 , 9398.274 , 10362.106 , 11335.749 , 12316.848 , 13310.895 , 14336.628 , 15428.775 , 16633.738 , 17998.21 , 19557.412 , 21331.355 , 23328.18 , 25550.145 , 27997.74 , 30670.965 , 33569.82 , 36694.305 , 40044.42 , 43620.164 , 47421.54 , 51448.547 , 55701.18 , 60179.445 ], dtype=float32) - hFacC(k, tile, j, i)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- vertical open fraction of tracer grid cell
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- Tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacC. The model allows for partially-filled cells to represent topographic variations more smoothly (hFacC < 1). Completely closed (dry) tracer grid cells have hFacC = 0. Note: the model z* coordinate system allows hFacC to vary through time. A time-invariant hFacC field is provided for reference.
array([[[[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ]], [[1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], ... [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - hFacW(k, tile, j, i_g)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- vertical open fraction of tracer grid cell 'west' face
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- The 'west' face of tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacW. The model allows for partially-filled cells for smoother representation of seafloor topography. Tracer grid cells adjacent in the 'x' direction that are partially closed in the vertical have hFacW < 1. The model z* coordinate system used by the model permits hFacC, and therefore hFacW, to vary through time. A time-invariant hFacW field is provided for reference. Note: The term 'west' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ]], [[1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], ... [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - hFacS(k, tile, j_g, i)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- vertical open fraction of tracer grid cell 'south' face
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- The 'south' face of tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacS. The model allows for partially-filled cells for smoother representation of seafloor topography. Tracer grid cells adjacent in the 'y' direction that are partially closed in the vertical have hFacS < 1. The model z* coordinate system used by the model permits hFacC, and therefore hFacS, to vary through time. A time-invariant hFacS field is provided for reference. Note: The term 'south' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
array([[[[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ]], [[1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], [1. , 1. , 1. , ..., 1. , 1. , 1. ], ... [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - maskC(k, tile, j, i)boolFalse False False ... False False
- long_name :
- wet/dry boolean mask for tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for tracer grid cells with nonzero open vertical fraction (hFacC > 0), otherwise False. Although hFacC can vary though time, cells will never close if starting open and will never open if starting closed: hFacC(i,j,k,t) > 0 for all t, if hFacC(i,j,k,t=0) and hFacC(i,j,k,t) = 0 for all t, if hFacC(i,j,k,t=0) = 0. Therefore, maskC is time invariant.
array([[[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True]], [[ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], ..., [ True, True, True, ..., False, True, True], [ True, True, True, ..., False, True, True], [ True, True, True, ..., False, False, True]], [[ True, True, True, ..., False, False, True], [ True, True, True, ..., True, False, True], [ True, True, True, ..., True, True, True], ..., ... ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]]) - maskW(k, tile, j, i_g)boolFalse False False ... False False
- long_name :
- wet/dry boolean mask for 'west' face of tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for grid cells with nonzero open vertical fraction along their 'west' face (hFacW > 0), otherwise False. Although hFacW can vary though time, cells will never close if starting open and will never open if starting closed: hFacW(i,j,k,t) > 0 for all t, if hFacW(i,j,k,t=0) and hFacW(i,j,k,t) = 0 for all t, if hFacW(i,j,k,t=0) = 0. Therefore, maskW is time invariant. Note:
array([[[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True]], [[ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], ..., [ True, True, True, ..., False, False, True], [ True, True, True, ..., False, False, True], [ True, True, True, ..., False, False, False]], [[ True, True, True, ..., False, False, False], [ True, True, True, ..., True, False, False], [ True, True, True, ..., True, True, True], ..., ... ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]]) - maskS(k, tile, j_g, i)boolFalse False False ... False False
- long_name :
- wet/dry boolean mask for 'south' face of tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for grid cells with nonzero open vertical fraction along their 'south' face (hFacS > 0), otherwise False. Although hFacS can vary though time, cells will never close if starting open and will never open if starting closed: hFacS(i,j,k,t) > 0 for all t, if hFacS(i,j,k,t=0) and hFacS(i,j,k,t) = 0 for all t, if hFacS(i,j,k,t=0) = 0. Therefore, maskS is time invariant. Note:
array([[[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True]], [[ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], [ True, True, True, ..., True, True, True], ..., [ True, True, True, ..., False, True, True], [ True, True, True, ..., False, True, True], [ True, True, True, ..., False, False, True]], [[ True, True, True, ..., False, False, True], [ True, True, True, ..., False, False, True], [ True, True, True, ..., True, False, True], ..., ... ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]]) - SSH(time, tile, j, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
array([[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.4789017 , -1.4837765 , -1.4820721 , ..., -1.2947402 , -1.2798777 , -1.2653834 ], [-1.4538441 , -1.4584619 , -1.455187 , ..., -1.2917751 , -1.2739644 , -1.255403 ], [-1.4215791 , -1.4232503 , -1.4176066 , ..., -1.290446 , -1.2703111 , -1.2480989 ]], [[-1.3808295 , -1.3784958 , -1.372 , ..., -1.2883928 , -1.2664716 , -1.2413179 ], [-1.3355613 , -1.3305387 , -1.3269446 , ..., -1.2839248 , -1.2608194 , -1.2335396 ], [-1.29667 , -1.2918196 , -1.2945448 , ..., -1.2746911 , -1.2511746 , -1.2229391 ], ... [ 0.02608651, 0.03203985, 0.05413032, ..., -1.2494164 , -1.2693772 , -1.2961768 ], [ 0.03012904, 0.0392129 , 0.06379545, ..., -1.2735504 , -1.295315 , -1.3238219 ], [ 0.03303316, 0.04432883, 0.07073619, ..., -1.290031 , -1.3149394 , -1.3470495 ]], [[-0.596936 , -0.6852058 , -0.769961 , ..., nan, nan, nan], [-0.58523434, -0.67554814, -0.76136804, ..., nan, nan, nan], [-0.5850948 , -0.67601234, -0.76148546, ..., nan, nan, nan], ..., [-1.325323 , -1.3553898 , -1.3883594 , ..., nan, nan, nan], [-1.3526964 , -1.3801088 , -1.4079852 , ..., nan, nan, nan], [-1.3783133 , -1.405505 , -1.4301533 , ..., nan, nan, nan]]]], dtype=float32) - SST(time, tile, j, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Potential temperature
- units :
- degree_C
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_potential_temperature
- comment :
- Sea water potential temperature is the temperature a parcel of sea water would have if moved adiabatically to sea level pressure. Note: the equation of state is a modified UNESCO formula by Jackett and McDougall (1995), which uses the model variable potential temperature as input assuming a horizontally and temporally constant pressure of $p_0=-g \rho_{0} z$.
- valid_min :
- -2.2909388542175293
- valid_max :
- 36.032955169677734
array([[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ 2.01509976e+00, 1.85003686e+00, 1.71582711e+00, ..., 1.26902759e+00, 1.27626359e+00, 1.28188074e+00], [ 2.16593409e+00, 2.02654552e+00, 1.91424274e+00, ..., 1.49355924e+00, 1.52569246e+00, 1.55790913e+00], [ 2.32527566e+00, 2.21751785e+00, 2.12773252e+00, ..., 1.66126490e+00, 1.71896994e+00, 1.78590453e+00]], [[ 2.48570871e+00, 2.40167022e+00, 2.32118011e+00, ..., 1.81008613e+00, 1.88386405e+00, 1.97170496e+00], [ 2.63559318e+00, 2.56559706e+00, 2.47433829e+00, ..., 1.95338202e+00, 2.03289223e+00, 2.12974238e+00], [ 2.75849938e+00, 2.69868946e+00, 2.57374191e+00, ..., 2.09234118e+00, 2.17186975e+00, 2.27002549e+00], ... 1.39164948e+00, 1.28957641e+00, 1.08142328e+00], [ 2.63058167e+01, 2.65797176e+01, 2.68715668e+01, ..., 1.24719834e+00, 1.07574773e+00, 7.48011649e-01], [ 2.63316193e+01, 2.65970135e+01, 2.68859653e+01, ..., 1.16007328e+00, 9.55231011e-01, 5.80619276e-01]], [[ 5.06779623e+00, 4.48910999e+00, 3.83241200e+00, ..., nan, nan, nan], [ 5.03983402e+00, 4.44997835e+00, 3.78785801e+00, ..., nan, nan, nan], [ 4.97232866e+00, 4.37776613e+00, 3.72398686e+00, ..., nan, nan, nan], ..., [ 7.23018527e-01, 2.29814187e-01, -2.17662126e-01, ..., nan, nan, nan], [ 2.79306203e-01, -2.17370406e-01, -6.34587169e-01, ..., nan, nan, nan], [ 5.89028262e-02, -4.86955911e-01, -9.32470202e-01, ..., nan, nan, nan]]]], dtype=float32) - SSS(time, tile, j, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Salinity
- units :
- 1e-3
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_salinity
- valid_min :
- 17.106637954711914
- valid_max :
- 41.26802444458008
- comment :
- Defined using CF convention 'Sea water salinity is the salt content of sea water, often on the Practical Salinity Scale of 1978. However, the unqualified term 'salinity' is generic and does not necessarily imply any particular method of calculation. The units of salinity are dimensionless and the units attribute should normally be given as 1e-3 or 0.001 i.e. parts per thousand.' see https://cfconventions.org/Data/cf-standard-names/73/build/cf-standard-name-table.html
array([[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [33.593304, 33.571117, 33.552166, ..., 33.94474 , 33.945957, 33.945374], [33.61394 , 33.597248, 33.585266, ..., 33.967583, 33.968716, 33.96788 ], [33.631084, 33.618633, 33.610626, ..., 33.976833, 33.97678 , 33.97494 ]], [[33.64739 , 33.638817, 33.63297 , ..., 33.978233, 33.977135, 33.974125], [33.664444, 33.661697, 33.65787 , ..., 33.97736 , 33.97548 , 33.97145 ], [33.68748 , 33.693874, 33.69012 , ..., 33.976746, 33.974003, 33.968967], ... [35.621944, 35.678062, 35.734776, ..., 33.776443, 33.713596, 33.610497], [35.60585 , 35.6631 , 35.72399 , ..., 33.7396 , 33.633568, 33.484848], [35.58173 , 35.633038, 35.698887, ..., 33.733326, 33.610363, 33.44627 ]], [[34.08766 , 34.054077, 34.017 , ..., nan, nan, nan], [34.087692, 34.053795, 34.016438, ..., nan, nan, nan], [34.0843 , 34.05046 , 34.01386 , ..., nan, nan, nan], ..., [33.457924, 33.276947, 33.146446, ..., nan, nan, nan], [33.315334, 33.179005, 33.087097, ..., nan, nan, nan], [33.272926, 33.144676, 33.08125 , ..., nan, nan, nan]]]], dtype=float32)
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2021-03-16T22:56:35
- date_issued :
- 2021-03-16T22:56:35
- date_metadata_modified :
- 2021-03-16T22:56:35
- date_modified :
- 2021-03-16T22:56:35
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- geospatial_vertical_max :
- 0.0
- geospatial_vertical_min :
- -6134.5
- geospatial_vertical_positive :
- up
- geospatial_vertical_resolution :
- variable
- geospatial_vertical_units :
- meter
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5A-GRD44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_GEOMETRY_LLC0090GRID_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- GRID_GEOMETRY_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides geometric parameters for the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Parameters include areas and lengths of grid cell sides, horizontal and vertical coordinates of grid cell centers and corners, grid rotation angles, and global domain geometry including bathymetry and land/ocean masks. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- title :
- ECCO Geometry Parameters for the Lat-Lon-Cap 90 (llc90) Native Model Grid (Version 4 Release 4)
- uuid :
- 87ff7d24-86e5-11eb-9c5f-f8f21e2ee3e0
Plotting a single tile with imshow¶
First we’ll plot the average SSH for the first month (Jan 2000) on tiles 3, 7, and 8 using the basic imshow routine from pyplot. We are plotting these three different tiles to show that these lat-lon-cap tiles all have a different orientation in  and
and  .
.
Note: Theorigin=’lower’argument to ``imshow`` is required to make the :math:`y` origin at the bottom of the plot.
Tile 2 (Northeast Atlantic)¶
[7]:
plt.figure(figsize=(6,5), dpi= 90)
# Step 1, select the tile to plot using the **.isel( )** syntax.
tile_to_plot = ecco_ds.SSH.isel(tile=2, time=0)
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=2,k=0) !=0, np.nan)
# Step 2, use plt.imshow()
plt.imshow(tile_to_plot, origin='lower');
# Step 3, add colorbar, title, and x and y axis labels
plt.colorbar()
plt.title('SSH (m) ' + str(ecco_ds.time[0].values)[0:7] + '\n tile 2')
plt.xlabel('x -->')
plt.ylabel('y -->')
[7]:
Text(0, 0.5, 'y -->')
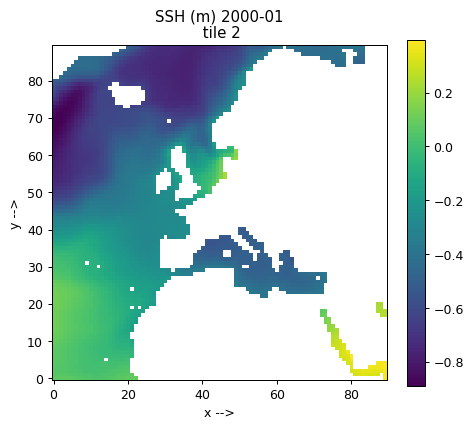
Tiles 0-5 are by default in a quasi-lat-lon orientation. +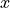 is to the east and +
is to the east and +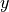 is to the north.
is to the north.
Tile 6 (the Arctic cap)¶
This time we’ll plot the Arctic cap tile 6. Notice the layout of the Arctic cap tile in 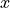 and
and  . We’ll follow the same procedure for plotting except we’ll use LaTeX to add arrows in the
. We’ll follow the same procedure for plotting except we’ll use LaTeX to add arrows in the  and
and  axis labels (for fun).
axis labels (for fun).
[8]:
plt.figure(figsize=(6,5), dpi= 90)
# Step 1, select the tile to plot using the **.isel( )** syntax.
tile_to_plot = ecco_ds.SSH.isel(tile=6, time=0)
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=6,k=0) !=0, np.nan)
# Step 2, use plt.imshow()
plt.imshow(tile_to_plot, origin='lower');
# Step 3, add colorbar, title, and x and y axis labels
plt.colorbar()
plt.title('SSH (m) ' + str(ecco_ds.time[0].values)[0:7] + '\n tile 6')
plt.xlabel('x -->');
plt.ylabel('y -->');
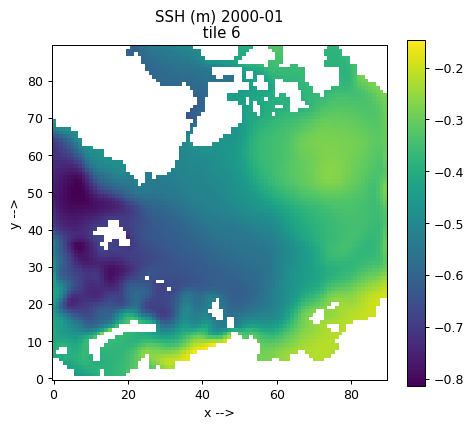
Because tile 6 is the Arctic cap, 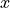 and
and 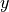 do not map to east and west throughout the domain.
do not map to east and west throughout the domain.
Tile 7 (N. Pacific / Bering Sea / Chukchi Sea)¶
For tiles 7-12 , positive 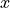 is southwards and positive
is southwards and positive 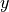 is eastwards.
is eastwards.
[9]:
plt.figure(figsize=(6,5), dpi= 90)
# pull out lats and lons
tile_num=7
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=1)
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, np.nan)
plt.imshow(tile_to_plot)
plt.colorbar()
plt.title('SSH (m) ' + str(ecco_ds.time[1].values)[0:7] + '\n tile ' + str(tile_num))
plt.xlabel('longitude');
plt.ylabel('latitude');
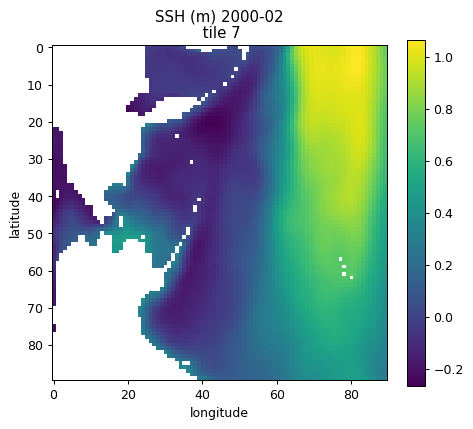
Tiles 7-12 are are also in a quasi-lat-lon orientation except that + is roughly south and +
is roughly south and + is roughly east.
is roughly east.
Plotting a single tile with pcolor and contourf¶
The pcolor and contourf routines allows us to add latitude and longitude to the figure. Because SSH is a ‘c’ point variable, its lat/lon coordinates are YC and XC
We can’t plot the Arctic cap tile with pcolor and contourf using latitude and longitude for the plot x and y axes because of the singularity at the pole and the 360 wrapping in longitude.
Instead, we will demonstrate pcolor and contourf for tile 2.
Tile 2 (Northeast N. Atlantic)¶
[10]:
fig=plt.figure(figsize=(10, 10))
tile_num=2
time_ind=1
# pull out lats and lons
lons = ecco_ds.XC.sel(tile=tile_num)
lats = ecco_ds.YC.sel(tile=tile_num)
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=time_ind)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, np.nan)
# create subplot for pcolor
fig = plt.subplot(221)
# use pcolor with 'lons' and 'lats' for the plot x and y axes
plt.pcolor(lons, lats, tile_to_plot, vmin=-1, vmax=1, cmap='RdBu_r')
plt.colorbar()
plt.title('PCOLOR SSH (m) \n' + str(ecco_ds.time[time_ind].values)[0:7] + '\n tile ' + str(tile_num))
plt.xlabel('longitude')
plt.ylabel('latitude')
# create subplot for contourf
fig=plt.subplot(222)
# use contourf with 'lons' and 'lats' for the plot x and y axes
plt.contourf(lons, lats, tile_to_plot, np.linspace(-1,1, 20,endpoint=True), cmap='RdBu_r', vmin=-1, vmax=1)
plt.title('CONTOURF SSH (m) \n' + str(ecco_ds.time[time_ind].values)[0:7] + '\n tile ' + str(tile_num))
plt.xlabel('longitude')
plt.ylabel('latitude')
plt.colorbar()
# plot every 3rd model grid line to show how tile 3 is 'warped' above around 60N
plt.plot(ecco_ds.XG.isel(tile=tile_num)[::3,::3], ecco_ds.YG.isel(tile=tile_num)[::3,::3],'k-')
plt.plot(ecco_ds.XG.isel(tile=tile_num)[::3,::3].T, ecco_ds.YG.isel(tile=tile_num)[::3,::3].T,'k-')
# push the subplots away from each other a bit
plt.subplots_adjust(bottom=0, right=1.2, top=.9)
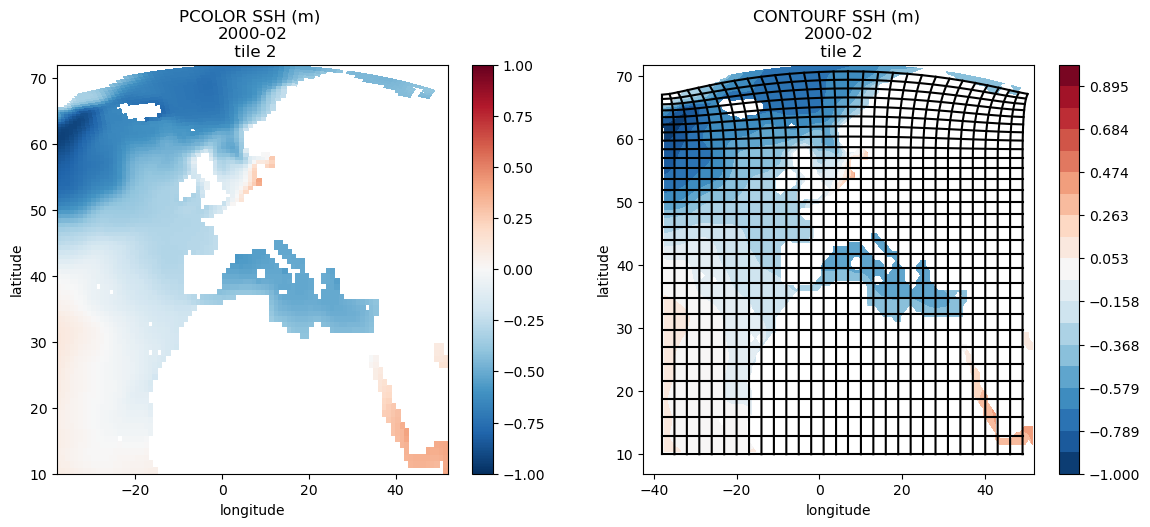
Tile 7 (N. Pacific / Bering Sea / Chukchi Sea)¶
If longitude and latitude are passed as the ‘x’ and ‘y’ arguments to pcolor and contourf then the fields will be oriented geographically.
[11]:
fig=plt.figure(figsize=(10, 10))
tile_num=7
time_ind=1
# pull out lats and lons
lons = np.copy(ecco_ds.XC.sel(tile=tile_num))
# we must convert the longitude coordinates from
# [-180 to 180] to [0 to 360]
# because of the crossing of the international date line.
lons[lons < 0] = lons[lons < 0]+360
lats = ecco_ds.YC.sel(tile=tile_num)
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=time_ind)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, np.nan)
# create subplot for pcolor
fig = plt.subplot(221)
# use pcolor with 'lons' and 'lats' for the plot x and y axes
plt.pcolor(lons, lats, tile_to_plot, vmin=-1, vmax=1.1, cmap='RdBu_r')
plt.colorbar()
plt.title('PCOLOR SSH (m) \n' + str(ecco_ds.time[time_ind].values)[0:7] + '\n tile ' + str(tile_num))
plt.xlabel('longitude')
plt.ylabel('latitude')
# create subplot for contourf
fig=plt.subplot(222)
# use contourf with 'lons' and 'lats' for the plot x and y axes
plt.contourf(lons, lats, tile_to_plot, np.linspace(-1,1.1,22,endpoint=True), cmap='RdBu_r', vmin=-1, vmax=1.1)
plt.title('CONTOURF SSH (m) \n' + str(ecco_ds.time[time_ind].values)[0:7] + '\n tile ' + str(tile_num))
plt.xlabel('longitude')
plt.ylabel('latitude')
plt.colorbar()
# push the subplots away from each other a bit
plt.subplots_adjust(bottom=0, right=1.2, top=.9)
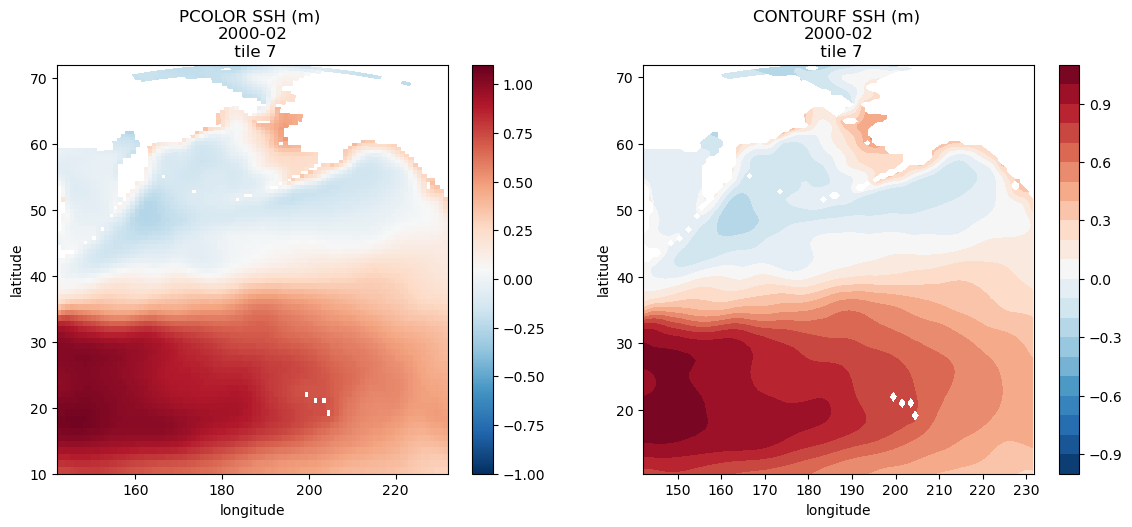
Plotting fields from one tile using Cartopy¶
The Cartopy package provides routines to make plots using different geographic projections. We’ll demonstrate plotting these three tiles again using Cartopy.
To see a list of Cartopy projections, see http://pelson.github.io/cartopy/crs/projections.html
Geographic Projections (AKA: plate carrée)¶
Cartopy works by transforming geographic coordintes (lat/lon) to new x,y coordinates associated with different projections. The most familiar projection is the so-called geographic projection (aka plate carree). When we plotted tiles using pcolor and contourf we were de-factor using the plate carree projection longitude and latitude were the ‘x’ and the ‘y’ of the plot.
With Cartopy we can make similar plots in the plate carree projection system and also apply some cool extra details, like land masks.
We’ll demonstrate on tiles 2 and 7 (again skipping tile 6 (Arctic cap) because we cannot use geographic coordinates as x and y when there is a polar singularity and 360 degrees of longitude.
Tile 2 with plate carree¶
[12]:
tile_num=2
time_ind=1
lons = ecco_ds.XC.isel(tile=tile_num)
lats = ecco_ds.YC.isel(tile=tile_num)
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=time_ind)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, np.nan)
fig = plt.figure(figsize=(10,5), dpi= 90)
# here is where you specify what projection you want to use
ax = plt.axes(projection=ccrs.PlateCarree())
# here is here you tell Cartopy that the projection
# of your 'x' and 'y' are geographic (lons and lats)
# and that you want to transform those lats and lons
# into 'x' and 'y' in the projection
cf = plt.contourf(lons, lats, tile_to_plot, 60,
transform=ccrs.PlateCarree());
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=2, color='gray', alpha=0.5, linestyle='--');
ax.coastlines()
ax.add_feature(cfeature.LAND)
# add separate axes for colorbar (to ensure it doesn't overlap main plot)
cbar_ax = fig.add_axes([0.88,0.1,0.02,0.8])
plt.colorbar(cf,ax=ax,cax=cbar_ax)
[12]:
<matplotlib.colorbar.Colorbar at 0x2926ca8f4f0>
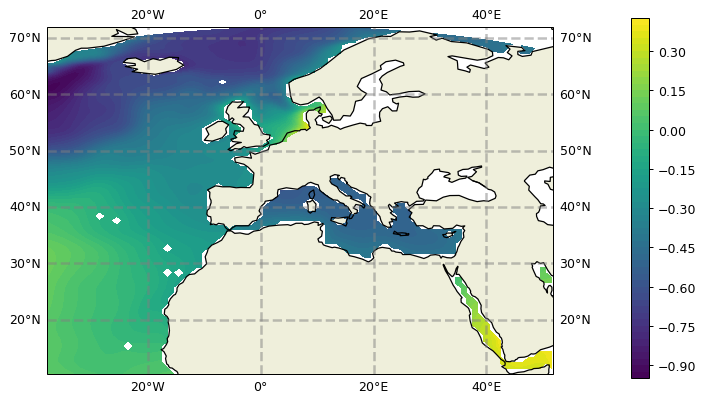
Other features we could have added include:
cartopy.feature.BORDERS
Country boundaries.
cartopy.feature.COASTLINE
Coastline, including major islands.
cartopy.feature.LAKES
Natural and artificial lakes.
cartopy.feature.LAND
Land polygons, including major islands.
cartopy.feature.OCEAN
Ocean polygons.
cartopy.feature.RIVERS
Single-line drainages, including lake centerlines.
Let’s add geographic borders just to demonstrate how extra features can be added to a Cartopy map
[13]:
fig = plt.figure(figsize=(10,5), dpi= 90)
# here is where you specify what projection you want to use
ax = plt.axes(projection=ccrs.PlateCarree())
# here is here you tell Cartopy that the projection
# of your 'x' and 'y' are geographic (lons and lats)
# and that you want to transform those lats and lons
# into 'x' and 'y' in the projection
cf = plt.contourf(lons, lats, tile_to_plot, 60,
transform=ccrs.PlateCarree());
gl = ax.gridlines(crs=ccrs.PlateCarree(), \
draw_labels=True,
linewidth=2, color='gray', \
alpha=0.5, linestyle='--');
ax.coastlines()
ax.add_feature(cfeature.LAND)
ax.add_feature(cfeature.BORDERS)
# add separate axes for colorbar (to ensure it doesn't overlap main plot)
cbar_ax = fig.add_axes([0.88,0.1,0.02,0.8])
plt.colorbar(cf,ax=ax,cax=cbar_ax)
[13]:
<matplotlib.colorbar.Colorbar at 0x2926cb8c940>
Tile 7 with plate carree¶
To use the plate carree projection across the international date line specify the central_longitude=-180 argument when defining the projection and for creating the gridlines. (see https://stackoverflow.com/questions/13856123/setting-up-a-map-which-crosses-the-dateline-in-cartopy)
[14]:
tile_num=7
time_ind=1
# pull out lats and lons
lons = np.copy(ecco_ds.XC.sel(tile=tile_num))
# we must convert the longitude coordinates from
# [-180 to 180] to [0 to 360]
# because of the crossing of the international date line.
lons[lons < 0] = lons[lons < 0]+360
lats = ecco_ds.YC.isel(tile=tile_num)
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=time_ind)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tile_to_plot= tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, np.nan)
fig = plt.figure(figsize=(10,5), dpi= 90)
# here is where you specify what projection you want to use
ax = plt.axes(projection=ccrs.PlateCarree(central_longitude=-180))
# here is here you tell Cartopy that the projection of your 'x' and 'y' are geographic (lons and lats)
# and that you want to transform those lats and lons into 'x' and 'y' in the projection
cf = plt.contourf(lons, lats, tile_to_plot, 60,
transform=ccrs.PlateCarree())
gl = ax.gridlines(crs=ccrs.PlateCarree(central_longitude=-180), draw_labels=True,
linewidth=2, color='gray', alpha=0.5, linestyle='--');
ax.coastlines()
ax.add_feature(cfeature.LAND)
# add separate axes for colorbar (to ensure it doesn't overlap main plot)
cbar_ax = fig.add_axes([0.88,0.1,0.02,0.8])
plt.colorbar(cf,ax=ax,cax=cbar_ax)
[14]:
<matplotlib.colorbar.Colorbar at 0x2925c6d9550>
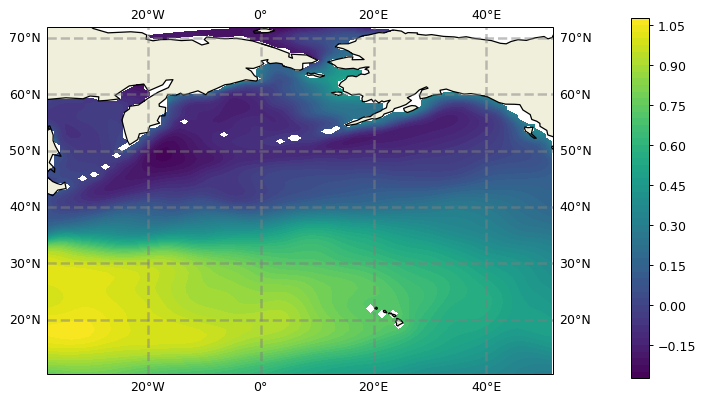
Polar Stereographic Projection¶
[15]:
tile_num=6
time_ind=1
# use lower-left grid corner coordinates for flat pcolormesh shading --
# this avoids some funky effects when plot is transformed into
# the polar stereographic projection
lon_corners = ecco_ds.XG.isel(tile=tile_num)
lat_corners = ecco_ds.YG.isel(tile=tile_num)
tile_to_plot = ecco_ds.SSH.isel(tile=tile_num, time=time_ind)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tile_to_plot = tile_to_plot.where(ecco_ds.hFacC.isel(tile=tile_num,k=0) !=0, \
np.nan)
plt.figure(figsize=(8,6), dpi= 90)
# Make a new projection, time of class "NorthPolarStereo"
ax = plt.axes(projection=ccrs.NorthPolarStereo(true_scale_latitude=70))
# here is here you tell Cartopy that the projection
# of your 'x' and 'y' are geographic (lons and lats)
# and that you want to transform those lats and lons
# into 'x' and 'y' in the projection
plt.pcolormesh(lon_corners, lat_corners, tile_to_plot[:-1,:-1],
transform=ccrs.PlateCarree(),shading='flat');
# plot land
ax.add_feature(cfeature.LAND)
ax.gridlines()
ax.coastlines()
plt.colorbar()
# Limit the map to 60 degrees latitude and above.
ax.set_extent([-180, 180, 60, 90], ccrs.PlateCarree())
# Compute a circle in axes coordinates, which we can use as a boundary
# for the map. We can pan/zoom as much as we like - the boundary will be
# permanently circular.
theta = np.linspace(0, 2*np.pi, 100)
center, radius = [0.5, 0.5], 0.5
verts = np.vstack([np.sin(theta), np.cos(theta)]).T
circle = mpath.Path(verts * radius + center)
#ax.set_boundary(circle, transform=ax.transAxes)
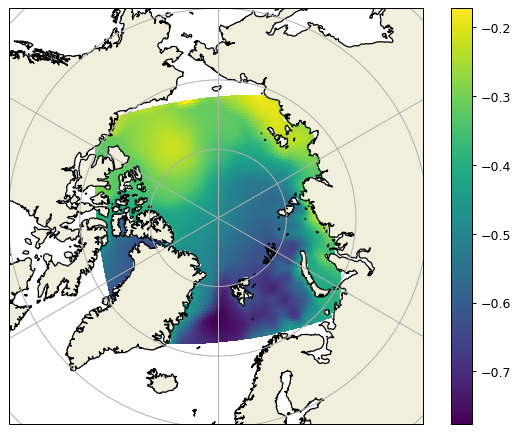
Plotting all 13 tiles simultaneously: No Projection¶
Plotting all 13 tiles with plot_tiles¶
The plot_tiles routine in the ecco_v4_py package makes plots of all 13 tiles of a field. By default the routine will plot all of the tiles in the lat-lon-cap layout shown earlier.
This routine will accept numpy arrays of dimension 13x90x90 or 2D slices of DataArrays with the same 13x90x90 dimension.
There are several additional arguments which we can access using the help command. Take a second to familiarize yourself with some of them.
[16]:
help(ecco.plot_tiles)
Help on function plot_tiles in module ecco_v4_py.tile_plot:
plot_tiles(tiles, cmap=None, layout='llc', rotate_to_latlon=False, Arctic_cap_tile_location=2, show_colorbar=False, show_cbar_label=False, show_tile_labels=True, cbar_label='', fig_size=9, less_output=True, **kwargs)
Plots the 13 tiles of the lat-lon-cap (LLC) grid
Parameters
----------
tiles : numpy.ndarray or dask.array.core.Array or xarray.core.dataarray.DataArray
an array of n=1..13 tiles of dimension n x llc x llc
- If *xarray DataArray* or *dask Array* tiles are accessed via *tiles.sel(tile=n)*
- If *numpy ndarray* tiles are acceed via [tile,:,:] and thus n must be 13.
cmap : matplotlib.colors.Colormap, optional
see plot_utils.assign_colormap for default
a colormap for the figure
layout : string, optional, default 'llc'
a code indicating the layout of the tiles
:llc: situates tiles in a fan-like manner which conveys how the tiles
are oriented in the model in terms of x an y
:latlon: situates tiles in a more geographically recognizable manner.
Note, this does not rotate tiles 7..12, it just places tiles
7..12 adjacent to tiles 0..5. To rotate tiles 7..12
specifiy *rotate_to_latlon* as True
rotate_to_latlon : boolean, default False
rotate tiles 7..12 so that columns correspond with
longitude and rows correspond to latitude. Note, this rotates
vector fields (vectors positive in x in tiles 7..12 will be -y
after rotation).
Arctic_cap_tile_location : int, default 2
integer, which lat-lon tile to place the Arctic tile over. can be
2, 5, 7 or 10.
show_colorbar : boolean, optional, default False
add a colorbar
show_cbar_label : boolean, optional, default False
add a label on the colorbar
show_tile_labels : boolean, optional, default True
show tiles numbers in subplot titles
cbar_label : str, optional, default '' (empty string)
the label to use for the colorbar
less_output : boolean, optional, default True
A debugging flag. True = less debugging output
cmin/cmax : floats, optional, default calculate using the min/max of the data
the minimum and maximum values to use for the colormap
fig_size : float, optional, default 9 inches
size of the figure in inches
fig_num : int, optional, default none
the figure number to make the plot in. By default make a new figure.
Returns
-------
f : matplotlib figure object
cur_arr : numpy ndarray
numpy array of size:
(llc*nrows, llc*ncols)
where llc is the size of tile for llc geometry (e.g. 90)
nrows, ncols refers to the subplot size
For now, only implemented for llc90, otherwise None is returned
We’ve seen this routine used in a few earlier tutorials. We’ll provide some additional examples below:
Default ‘native grid’ layout¶
[17]:
# optional arguments:
# cbar - show the colorbar
# cmin, cmax - color range min and max
# fsize - figure size in inches
# pull out surface salinity
tmp_plt = ecco_ds.SSS.isel(time=3)
# mask to NaN where hFacC is == 0
# syntax is actually "keep where hFacC is not equal to zero"
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) != 0, np.nan)
[18]:
ecco.plot_tiles(tmp_plt, \
cmin=32, \
cmax=35.5, \
cmap='RdYlBu_r', \
show_colorbar=True);
# use `suptitle` (super title) to make a title over subplots.
plt.suptitle('SSS ' + str(ecco_ds.time[3].values)[0:7]);
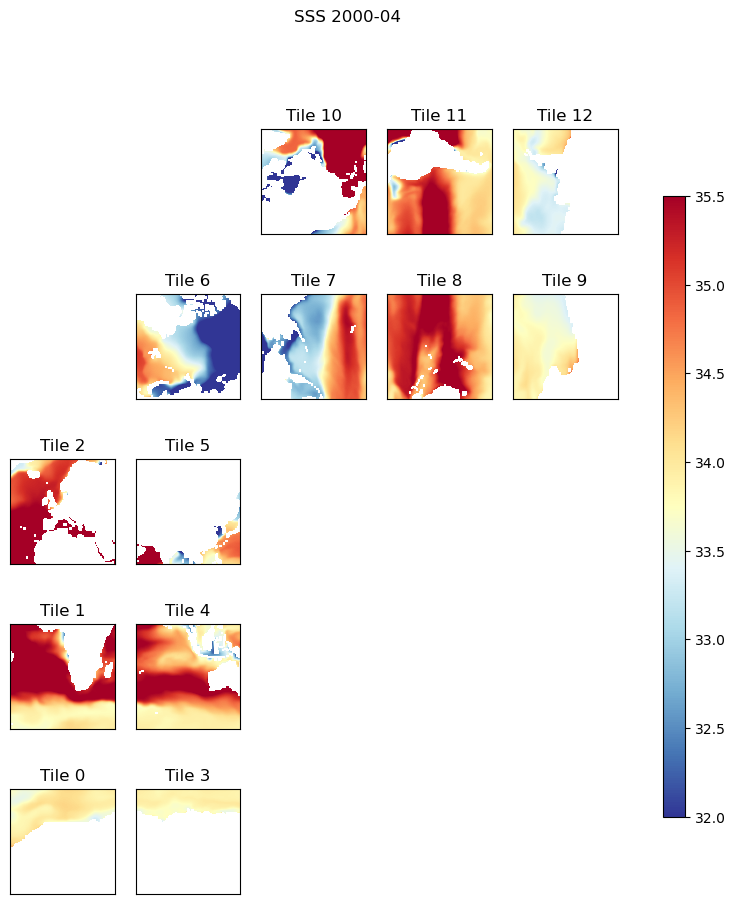
lat-lon layout¶
Another option of plot_tiles is to show tiles 7-12 rotated and lined up tiles 0-5
Note: Rotation of tiles 7-13 is only forplotting. These arrays are not rotated using this routine. We’ll show to how actually rotate these tiles in a later tutorial.
[19]:
# optional arguments:
# cbar - show the colorbar
# cmin, cmax - color range min and max
# fsize - figure size in inches
tmp_plt = ecco_ds.SSS.isel(time=3)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) != 0, np.nan)
ecco.plot_tiles(tmp_plt, \
cmin=32, cmax=35.5, cmap='RdYlBu_r', \
show_colorbar=True, fig_size=8,\
layout='latlon', \
rotate_to_latlon=True);
# use `suptitle` (super title) to make a title over subplots.
plt.suptitle('SSS ' + str(ecco_ds.time[3].values)[0:7]);
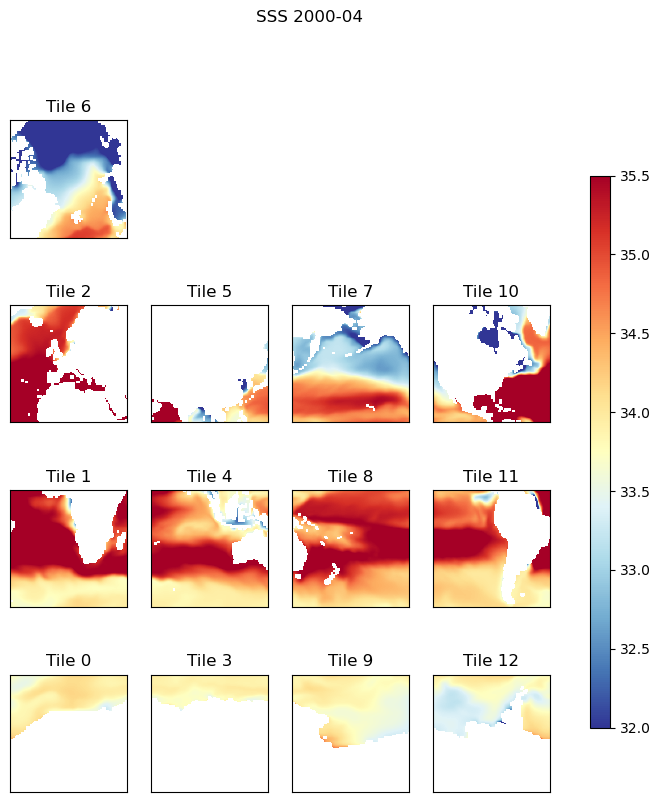
The version of plot_tiles is to remove the tile labels and put the titles together in a tight formation and sticks the Arctic tile over tile 10
[20]:
# optional arguments:
# cbar - show the colorbar
# cmin, cmax - color range min and max
# fsize - figure size in inches
tmp_plt = ecco_ds.SSS.isel(time=3)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) != 0, np.nan)
ecco.plot_tiles(tmp_plt, cmin=32, cmax=35.5, cmap='RdYlBu_r', \
show_colorbar=True, fig_size=8,\
layout='latlon',rotate_to_latlon=True,\
show_tile_labels=False, \
Arctic_cap_tile_location=10)
# use `suptitle` (super title) to make a title over subplots.
plt.suptitle('SSS ' + str(ecco_ds.time[3].values)[0:7]);
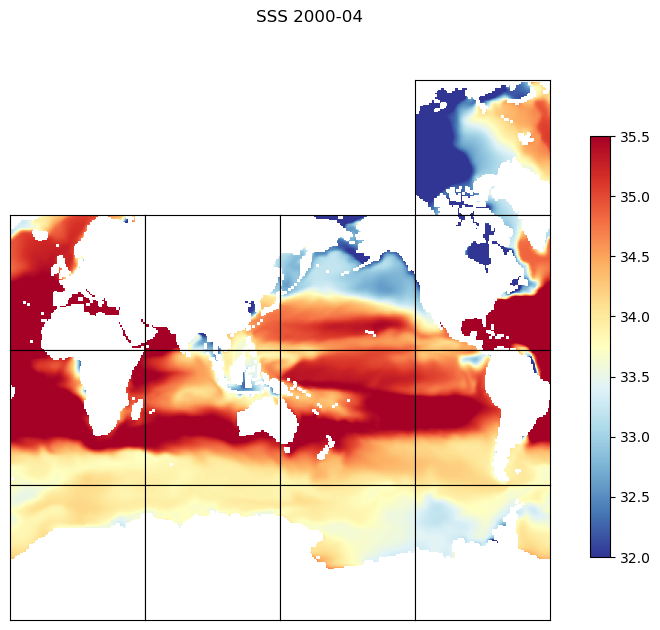
Almost ready for the hyperwall!
You can plot a subset of tiles using plot_tiles, but you need to pass it a full xarray DataArray or Numpy array with 13 tiles, and the undesired tiles masked out.
[21]:
# optional arguments:
# cbar - show the colorbar
# cmin, cmax - color range min and max
# fsize - figure size in inches
tmp_plt = ecco_ds.SSS.isel(time=3)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) != 0, np.nan)
tiles_to_subset = [1,2,4,5]
# add dimensions to vector of tile numbers, so it will readily broadcast across grid
tile_to_broadcast = np.expand_dims(ecco_ds.tile.values,axis=(-2,-1))
# mask tiles not in tiles_to_subset with NaNs
tmp_plt_subset = tmp_plt.where(np.isin(tile_to_broadcast,tiles_to_subset),np.nan)
# select a subset of tiles
ecco.plot_tiles(tmp_plt_subset, cmin=32, cmax=35.5, \
cmap='RdYlBu_r', show_colorbar=True, fig_size=8,\
layout='latlon',rotate_to_latlon=True,\
show_tile_labels=False)
# use `suptitle` (super title) to make a title over subplots.
plt.suptitle('SSS ' + str(ecco_ds.time[3].values)[0:7]);
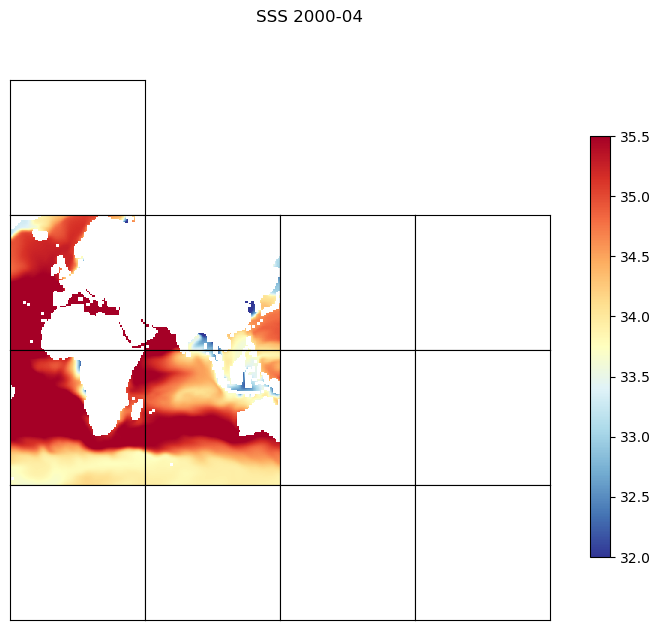
Plotting all 13 tiles with plot_proj_to_latlon_grid¶
Our routine plot_proj_to_latlon_grid takes numpy arrays or DataArrays with 13 tiles and creates global plots with one of three types of projections (passed as arguments to the function): ~~~ projection_type : string, optional denote the type of projection, options include ‘robin’ - Robinson ‘PlateCaree’ - flat 2D projection ‘Mercator’ ‘cyl’ - Lambert Cylindrical ‘ortho’ - Orthographic ‘stereo’ - polar stereographic projection, see lat_lim for choosing ‘InterruptedGoodeHomolosi ~~~
Before plotting this routine interpolates the the filed onto a lat-lon grid (default resoution 0.25 degree) to conform with Cartopy's requirement that the fields to be transformed be on regular square grid.
There are only three argument required of plot_proj_to_latlon_grid, an array of longitudes, an array of latitudes, and an array of the field you wish to plot. The arrays can be either numpy arrays or DataArrays.
Let’s again spend a second to look at the optional arguments available to us in this routine:
[22]:
help(ecco.plot_proj_to_latlon_grid)
Help on function plot_proj_to_latlon_grid in module ecco_v4_py.tile_plot_proj:
plot_proj_to_latlon_grid(lons, lats, data, projection_type='robin', dx=0.25, dy=0.25, radius_of_influence=112000, plot_type='pcolormesh', cmap=None, cmin=None, cmax=None, user_lon_0=0, user_lat_0=None, lat_lim=50, parallels=None, show_coastline=True, show_colorbar=False, show_land=True, show_grid_lines=True, show_grid_labels=False, show_coastline_over_data=True, show_land_over_data=True, grid_linewidth=1, grid_linestyle='--', colorbar_label=None, subplot_grid=None, less_output=True, **kwargs)
Plot a field of data from an arbitrary grid with lat/lon coordinates
on a geographic projection after resampling it to a regular lat/lon grid.
Parameters
----------
lons, lats : numpy ndarray or xarray DataArrays, required
the longitudes and latitudes of the data to plot
data : numpy ndarray or xarray DataArray, required
the field to be plotted
dx, dy : float, optional, default 0.25 degrees
latitude, longitude spacing of the new lat/lon grid onto which the
field 'data' will be resampled.
radius_of_influence : float, optional, default 112000 m
to map values from 'data' to the new lat/lon grid, we use use a
nearest neighbor approach with the constraint that we only use values
from 'data' that fall within a circle with radius='radius_of_influence'
from the center of each new lat/lon grid cell.
for the llc90, with 1 degree resolution,
radius_of_influence = 1/2 x sqrt(2) x 112e3 km
would suffice.
projection_type : string, optional
denote the type of projection, see Cartopy docs.
options include
'robin' - Robinson
'PlateCarree' - flat 2D projection
'LambertConformal'
'Mercator'
'EqualEarth'
'Mollweide'
'AlbersEqualArea'
'cyl' - Lambert Cylindrical
'ortho' - Orthographic
'stereo' - polar stereographic projection, see lat_lim for choosing
'InterruptedGoodeHomolosine'
North or South
plot_type : string, optional
denotes type of plot ot make with the data
options include
'pcolormesh' - pcolormesh
'contourf' - filled contour
'points' - plot points at lat/lon locations
cmap : matplotlib.colors.Colormap, optional, default None
a colormap for the figure.
cmin/cmax : floats, optional, default None
the minimum and maximum values to use for the colormap
if not specified, use the full range of the data
user_lon_0 : float, optional, default 0 degrees
denote central longitude
user_lat_0 : float, optional, default None
denote central latitude (for relevant projections only, see Cartopy)
lat_lim : int, optional, default 50 degrees
for stereographic projection, denote the Southern (Northern) bounds for
North (South) polar projection or cutoff for LambertConformal projection
parallels : float, optional,
standard_parallels, one or two latitudes of correct scale
(for relevant projections only, see Cartopy docs)
show_coastline : logical, optional, default True
show coastline or not
show_colorbar : logical, optional, default False
show a colorbar or not,
show_land : logical, optional, default True
show land or not
show_grid_lines : logical, optional, default True
True only possible for some cartopy projections
show_grid_labels: logical, optional, default False
True only possible for some cartopy projections
show_coastline_over_data : logical, optional, default True
draw coastline over the data or under the data
show_land_over_data: logical, optional, default True
draw land over the data or under the data
grid_linewidth : float, optional, default 1.0
width of grid lines
grid_linestyle : string, optional, default = '--'
pattern of grid lines,
subplot_grid : dict or list, optional
specifying placement on subplot as
dict:
{'nrows': rows_val, 'ncols': cols_val, 'index': index_val}
list:
[nrows_val, ncols_val, index_val]
equates to
matplotlib.pyplot.subplot(
row=nrows_val, col=ncols_val,index=index_val)
less_output : string, optional
debugging flag, don't print if True
plot_proj_to_latlon_grid(lons, lats, data, projection_type=’robin’, plot_type=’pcolormesh’, user_lon_0=-66, lat_lim=50, levels=20, cmap=’jet’, dx=0.25, dy=0.25, show_colorbar=False, show_grid_lines=True, show_grid_labels=True, subplot_grid=None, less_output=True, **kwargs)
Robinson projection¶
First we’ll demonstrate the Robinson projection interpolated to a 2x2 degree grid
[23]:
plt.figure(figsize=(12,6), dpi= 90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC, \
ecco_ds.YC, \
tmp_plt, \
plot_type = 'pcolormesh', \
dx=2,\
dy=2, \
projection_type = 'robin',\
less_output = False);
_create_projection_axis: projection_type robin
_create_projection_axis: user_lon_0, user_lat_0 0 None
_create_projection_axis: parallels None
_create_projection_axis: lat_lim 50
Projection type: robin
-179.0 179.0
-180.0 180.0
-89.0 89.0
-90.0 90.0
Setting lon_0 = 110 or -66 yield a global centering that is is more usesful for plotting ocean basins.
[24]:
plt.figure(figsize=(12,6), dpi= 90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC,
ecco_ds.YC,
tmp_plt,user_lon_0=-66,
plot_type = 'pcolormesh', dx=2,dy=2);
-179.0 113.0
-180.0 114.0
-89.0 89.0
-90.0 90.0
115.03125984375001 178.96875015625
114.00001 180.0
-89.0 89.0
-90.0 90.0
Cylindrical projection¶
Try the Cylindrical Projection with an interpolated lat-lon resolution of 0.25 degrees and pcolormesh.
[25]:
plt.figure(figsize=(12,6), dpi= 90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC, ecco_ds.YC, \
tmp_plt, \
user_lon_0=-66,\
projection_type='cyl',\
plot_type = 'pcolormesh', \
dx=.25,dy=.25);
-179.875 113.875
-180.0 114.0
-89.875 89.875
-90.0 90.0
114.1254852661597 179.8745247338403
114.00001 180.0
-89.875 89.875
-90.0 90.0
Polar stereographic projection¶
Another projection built into plot_proj_to_latlon_grid is polar stereographic. The argument lat_lim determines the limit of this type of projection. If lat_lim is postive, the projection is centered around the north pole and vice versa.
Northern Hemisphere¶
[26]:
plt.figure(figsize=(12,6), dpi= 90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC, ecco_ds.YC, \
tmp_plt, \
projection_type='stereo',\
plot_type = 'contourf', \
show_colorbar=True,
dx=1, dy=1,cmin=-1, cmax=1,\
lat_lim=40);
-179.5 179.5
-180.0 180.0
-89.5 89.5
-90.0 90.0
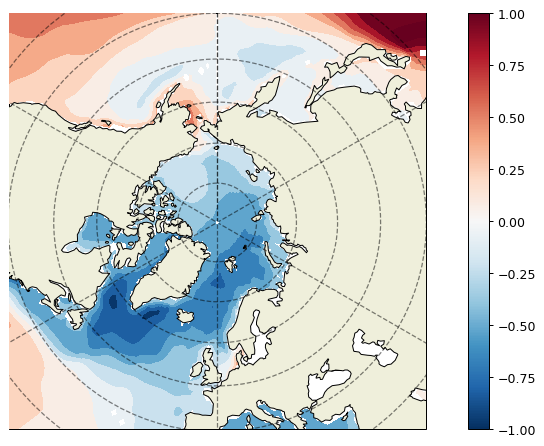
Southern Hemisphere¶
The final example is a south-pole centered plot. Note that lat_lim is now negative.
[27]:
plt.figure(figsize=(12,6), dpi= 90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC, ecco_ds.YC, \
tmp_plt, \
projection_type='stereo',\
plot_type = 'contourf', \
show_colorbar=True,
dx=1, dy=1,\
lat_lim=-40,cmin=-2,cmax=2);
-179.5 179.5
-180.0 180.0
-89.5 89.5
-90.0 90.0
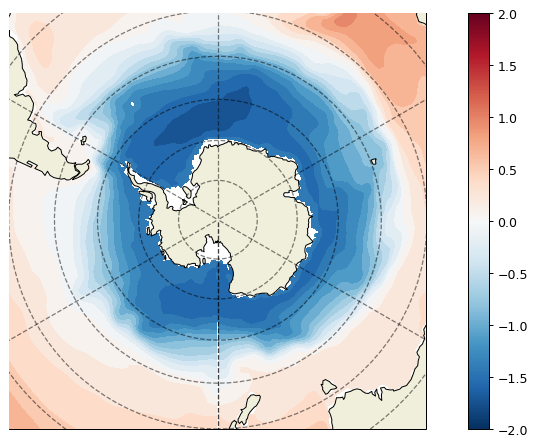
Conclusion¶
You now know several ways of plotting ECCO state estimate fields. There is a lot more to explore with Cartopy - dive in and start making your own cool plots!
[28]:
plt.figure(figsize=(16,6), dpi=90)
tmp_plt = ecco_ds.SSH.isel(time=1)
tmp_plt = tmp_plt.where(ecco_ds.hFacC.isel(k=0) !=0)
ecco.plot_proj_to_latlon_grid(ecco_ds.XC, ecco_ds.YC, \
tmp_plt, \
user_lon_0=-66,\
projection_type='InterruptedGoodeHomolosine',\
plot_type = 'pcolormesh', \
show_colorbar=True,
dx=1, dy=1);
plt.title('ECCO SSH [m] -- like never before! :)');
-179.5 113.5
-180.0 114.0
-89.5 89.5
-90.0 90.0
114.50770223076924 179.49230776923076
114.00001 180.0
-89.5 89.5
-90.0 90.0