ECCOv4 Loading llc binary files in the ‘compact’ format¶
Objective¶
To teach how to loading ECCO binary files written by the MITgcm in the llc ‘compact’ format.
Introduciton¶
When the MITgcm saves diagnostic and other fields to files it does so using the so-called ‘compact’ format. The compact format distributes the arrays from the 13 lat-lon-cap tiles in a somewhat unintuitive manner. Fortunately, it is not difficult to extract the 13 tiles from ‘compact’ format files. This tutorial will show you how to use the ‘read_llc_to_tiles’ subroutine to read and re-organize MITgcm’s files written in compact format into a more familiar 13-tile layout.
Objectives¶
By the end of the tutorial you will be able to read llc compact binary files of any dimension, plot them, and convert them into DataArrays.
[1]:
## Import the ecco_v4_py library into Python
## =========================================
## -- If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. For example, if your ecco_v4_py
## files are in /Users/ifenty/ECCOv4-py/ecco_v4_py, then use:
import sys
sys.path.append('/Users/ifenty/git_repos/my_forks/ECCOv4-py')
import ecco_v4_py as ecco
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
The read_llc_to_tiles subroutine¶
read_llc_to_tiles reads a llc compact format binary file and converts to a numpy ndarray of dimension: [N_recs, N_z, N_tiles, llc, llc]
For ECCOv4 our convention is:
'N_recs' = number of time levels
'N_z' = number of depth levels
'N_tiles' = 13
'llc' = 90
By default the routine will try to load a single 2D slice of a llc90 compact binary file: (N_recs = 1, N_z =1, N_tiles = 13, and llc=90).
There are several other options which you can learn about using the ‘help’ command:
[2]:
help(ecco.read_llc_to_tiles)
Help on function read_llc_to_tiles in module ecco_v4_py.read_bin_llc:
read_llc_to_tiles(fdir, fname, llc=90, skip=0, nk=1, nl=1, filetype='>f', less_output=False, use_xmitgcm=False)
Loads an MITgcm binary file in the 'tiled' format of the
lat-lon-cap (LLC) grids with dimension order:
[N_recs, N_z, N_tiles, llc, llc]
where if either N_z or N_recs =1, then that dimension is collapsed
and not present in the returned array.
if use_xmitgcm == True
data are read in via the low level routine
xmitgcm.utils.read_3d_llc_data and returned as dask array.
Hint: use data_tiles.compute() to load into memory.
if use_xmitgcm == False
Loads an MITgcm binary file in the 'compact' format of the
lat-lon-cap (LLC) grids and converts it to the '13 tiles' format
of the LLC grids.
Parameters
----------
fdir : string
A string with the directory of the binary file to open
fname : string
A string with the name of the binary file to open
llc : int
the size of the llc grid. For ECCO v4, we use the llc90 domain
so `llc` would be `90`.
Default: 90
skip : int
the number of 2D slices (or records) to skip.
Records could be vertical levels of a 3D field, or different 2D fields, or both.
nk : int
number of 2D slices (or records) to load in the third dimension.
if nk = -1, load all 2D slices
Default: 1 [singleton]
nl : int
number of 2D slices (or records) to load in the fourth dimension.
Default: 1 [singleton]
filetype: string
the file type, default is big endian (>) 32 bit float (f)
alternatively, ('<d') would be little endian (<) 64 bit float (d)
less_output : boolean
A debugging flag. False = less debugging output
Default: False
use_xmitgcm : boolean
option to use the routine xmitgcm.utils.read_3d_llc_data into a dask
array, i.e. not into memory.
Otherwise read in as a compact array, convert to faces, then to tiled format
Default: False
Returns
-------
data_tiles
a numpy array of dimension 13 x nl x nk x llc x llc, one llc x llc array
for each of the 13 tiles and nl and nk levels.
Related routines¶
Two related routines which you might find useful:
read_llc_to_compact: Loads an MITgcm binary file in the ‘compact’ format of the lat-lon-cap (LLC) grids and preserves its original dimension
read_llc_to_faces : Loads an MITgcm binary file in the ‘compact’ format of the lat-lon-cap (LLC) grids and converts it to the ‘5 faces’ dictionary.
For the remainder of the tutorial we will only use read_llc_to_tiles.
Example 1: Load a 2D llc ‘compact’ binary file¶
The file ‘bathy_eccollc_90x50_min2pts.bin’ contains the 2D array of bathymetry for the model.
[3]:
input_dir = '/Users/ifenty/tmp/input_init/'
input_file = 'bathy_eccollc_90x50_min2pts.bin'
read_llc_to_tiles actually runs several other subroutines: load_binary_array which does the lower level reading of the binary file, llc_compact_to_faces which converts the array to 5 ‘faces’, and finally llc_faces_to_tiles which extracts the 13 tiles from the 5 faces:
[4]:
bathy = ecco.read_llc_to_tiles(input_dir, input_file)
load_binary_array: loading file /Users/ifenty/tmp/input_init/bathy_eccollc_90x50_min2pts.bin
load_binary_array: data array shape (1170, 90)
load_binary_array: data array type >f4
llc_compact_to_faces: dims, llc (1170, 90) 90
llc_compact_to_faces: data_compact array type >f4
llc_faces_to_tiles: data_tiles shape (13, 90, 90)
llc_faces_to_tiles: data_tiles dtype >f4
bathy is a numpy float32 array with dimension [13, 90, 90]
Plot the 13 tiles bathymetry data¶
[5]:
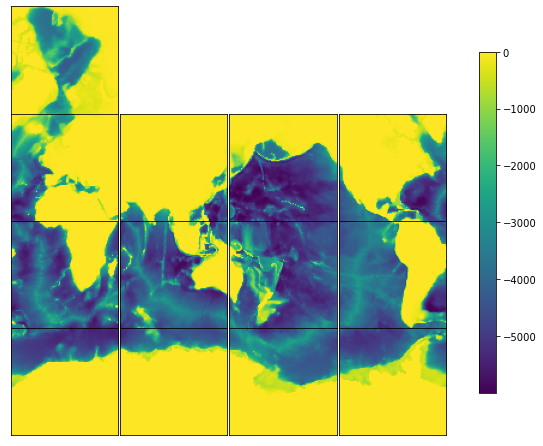
# Use plot_tiles to make a quick plot of the 13 tiles. See the tutorial on plotting for more examples.
ecco.plot_tiles(bathy, layout='latlon',rotate_to_latlon=True,show_tile_labels=False, show_colorbar=True);
Load ecco-grid information to make a fancier lat-lon plot¶
With the longitudes (XC) and latitudes (YC) and the 13 tile ndarray we can plot the field in different geographic projections. See the tutorial on plotting for more examples.
[6]:
ecco_grid_dir = '/Users/ifenty/tmp/nctiles_grid/'
ecco_grid = ecco.load_ecco_grid_nc(input_dir, 'ECCO-GRID.nc')
[7]:
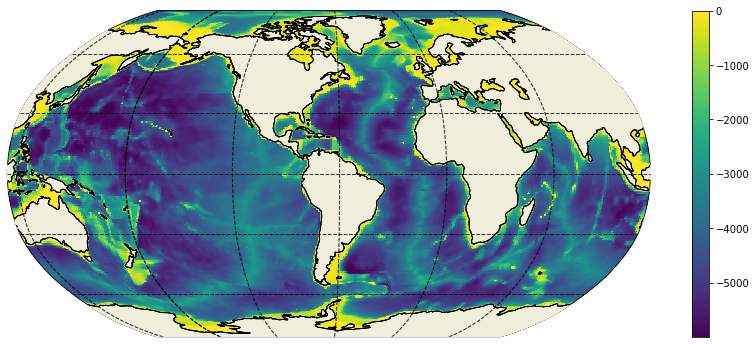
plt.figure(figsize=(15,6));
ecco.plot_proj_to_latlon_grid(ecco_grid.XC, ecco_grid.YC, bathy, show_colorbar=True, user_lon_0=-66);
[8]:
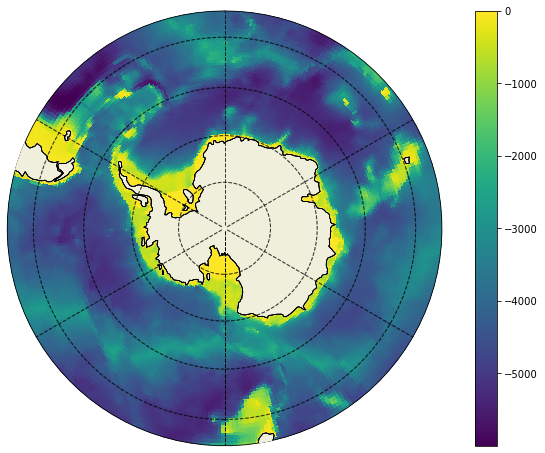
plt.figure(figsize=(12,8));
ecco.plot_proj_to_latlon_grid(ecco_grid.XC, ecco_grid.YC, bathy, show_colorbar=True,\
projection_type='stereo', lat_lim =-45, less_output=True,dx=.25,dy=.25);
Convert the ndarray into a DataArray¶
Converting the ndarray to a DataArray can be useful for broadcasting operations. One approach is to create the DataArray manually by specifying the names and values of the new dimension coordinates:
[9]:
tile = range(13)
i = range(90)
j = range(90)
[10]:
# Convert numpy array to an xarray DataArray with matching dimensions as the monthly mean fields
bathy_DA = xr.DataArray(bathy,coords={'tile': tile,
'j': j,
'i': i}, dims=['tile','j','i'])
[11]:
print(bathy_DA.dims)
('tile', 'j', 'i')
Another approach is to use the routine llc_tiles_to_xda. llc_tiles_to_xda uses
[12]:
bathy.shape
[12]:
(13, 90, 90)
[13]:
bathy_DA2 = ecco.llc_tiles_to_xda(bathy, var_type='c',grid_da=ecco_grid.XC)
print(bathy_DA2.dims)
print(bathy_DA2.coords)
('tile', 'j', 'i')
Coordinates:
* j (j) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int64 0 1 2 3 4 5 6 7 8 9 10 11 12
XC (tile, j, i) float32 -111.60647 -111.303 ... -105.58465 -111.86579
YC (tile, j, i) float32 -88.24259 -88.382515 ... -88.07871 -88.10267
CS (tile, j, i) float32 0.06157813 0.06675376 ... -0.9983638
SN (tile, j, i) float32 -0.99810225 -0.9977695 ... -0.057182025
Depth (tile, j, i) float32 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
rA (tile, j, i) float32 362256450.0 363300960.0 ... 361119100.0
Example 2: Load a 3D ‘compact’ llc binary file with 3rd dimension = Time¶
The file ‘runoff-2d-Fekete-1deg-mon-V4-SMOOTH.bin’ contains the 12 month climatology of river runoff, dimensions of [time, j, i].
[14]:
input_file = 'runoff-2d-Fekete-1deg-mon-V4-SMOOTH.bin'
specify the length of the n_recs dimension, nl, as 12. By default, nk = 1
[15]:
runoff = ecco.read_llc_to_tiles(input_dir, input_file, nl=12)
load_binary_array: loading file /Users/ifenty/tmp/input_init/runoff-2d-Fekete-1deg-mon-V4-SMOOTH.bin
load_binary_array: data array shape (12, 1, 1170, 90)
load_binary_array: data array type >f4
llc_compact_to_faces: dims, llc (12, 1, 1170, 90) 90
llc_compact_to_faces: data_compact array type >f4
llc_faces_to_tiles: data_tiles shape (12, 1, 13, 90, 90)
llc_faces_to_tiles: data_tiles dtype >f4
Shape is n_recs (12), n_z (1), n_tiles (13), llc (90), llc (90)
[16]:
print(runoff.shape)
(12, 1, 13, 90, 90)
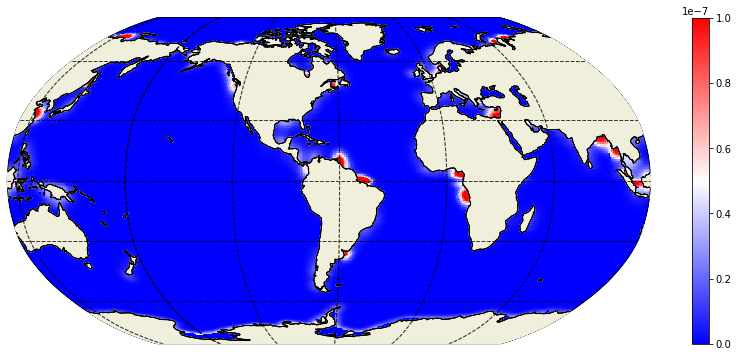
Plot the November runoff climatology¶
[17]:
plt.figure(figsize=(15,6));
ecco.plot_proj_to_latlon_grid(ecco_grid.XC, ecco_grid.YC, runoff[10,:], cmin=0,cmax=1e-7,\
show_colorbar=True, cmap='bwr',user_lon_0=-66);
Convert the ndarray into a DataArray¶
Converting the ndarray to a DataArray can be useful for broadcasting operations. Two methods: ‘manual’ and using llc_tiles_to_xda.
Method 1: Manual¶
[18]:
tile = range(1,14)
i = range(90)
j = range(90)
time = range(12) # months
k = [0];
[19]:
# Convert numpy array to an xarray DataArray with matching dimensions as the monthly mean fields
runoff_DA = xr.DataArray(runoff,coords={'time': time,
'k': k,
'tile': tile,
'j': j,
'i': i}, dims=['time','k','tile','j','i'])
[20]:
print(runoff_DA.dims)
print(runoff_DA.shape)
('time', 'k', 'tile', 'j', 'i')
(12, 1, 13, 90, 90)
Method 2: llc_tiles_to_xda¶
runoff is a 3D array with the 3rd dimension being time and therefore we need to pass llc_tiles_to_xda a similarly-dimensioned DataArray or tell the subroutine that the 3rd dimension is depth. The ‘ecco_grid’ DataSet doesn’t have any similary-dimensioned DataArrays (no DataArrays with a time dimension). Therefore we will tell the routine that the new 4th dimension should be time:
[21]:
##### specify that the 5th dimension should be time
runoff_DA2 = ecco.llc_tiles_to_xda(runoff, var_type='c',dim4='depth', dim5='time')
print(runoff_DA2.dims)
print(runoff_DA2.coords)
('time', 'k', 'tile', 'j', 'i')
Coordinates:
* k (k) int64 0
* time (time) int64 0 1 2 3 4 5 6 7 8 9 10 11
* tile (tile) int64 0 1 2 3 4 5 6 7 8 9 10 11 12
* j (j) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
Example 3: Load a 3D ‘compact’ llc binary file with 3rd dimension = Depth¶
The file ‘total_kapredi_r009bit11.bin’ is a 50 depth level array of the adjusted Redi parameter from Release 1 (first guess + adjustments), dimensions of [depth, j, i]. Release 4’s adjusted Redi parameter can be found at https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/ECCO_L4_OCEAN_3D_MIX_COEFFS_LLC0090GRID_V4R4/OCEAN_3D_MIXING_COEFFS_ECCO_V4r4_native_llc0090.nc.
[22]:
input_file = 'total_kapredi_r009bit11.bin'
specify the number of depth levels as 50. n_recs defaults to 1 and is dropped by default.
[23]:
kapredi = ecco.read_llc_to_tiles(input_dir, input_file, nk=50)
load_binary_array: loading file /Users/ifenty/tmp/input_init/total_kapredi_r009bit11.bin
load_binary_array: data array shape (50, 1170, 90)
load_binary_array: data array type >f4
llc_compact_to_faces: dims, llc (50, 1170, 90) 90
llc_compact_to_faces: data_compact array type >f4
llc_faces_to_tiles: data_tiles shape (50, 13, 90, 90)
llc_faces_to_tiles: data_tiles dtype >f4
[24]:
print(kapredi.shape)
(50, 13, 90, 90)
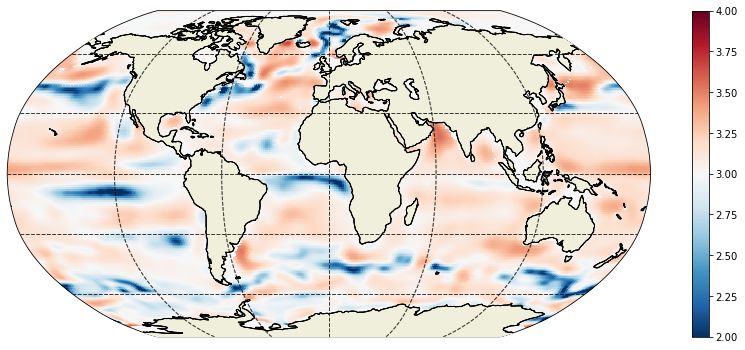
Plot log10 of the parameter at the 10th depth level (105m)¶
[25]:
plt.figure(figsize=(15,6));
ecco.plot_proj_to_latlon_grid(ecco_grid.XC, ecco_grid.YC, np.log10(kapredi[10,:]),\
cmin=2,cmax=4,show_colorbar=True);
<ipython-input-25-b09b1aa1ac7e>:2: RuntimeWarning: divide by zero encountered in log10
ecco.plot_proj_to_latlon_grid(ecco_grid.XC, ecco_grid.YC, np.log10(kapredi[10,:]),\
Convert the ndarray into a DataArray¶
Converting the ndarray to a DataArray can be useful for broadcasting operations. Two methods: ‘manual’ and using llc_tiles_to_xda.
Method 1: Manual¶
[26]:
tile = range(1,14)
i = range(90)
j = range(90)
k = range(50)
[27]:
# Convert numpy array to an xarray DataArray with matching dimensions as the monthly mean fields
kapredi_DA = xr.DataArray(kapredi,coords={'k': k,
'tile': tile,
'j': j,
'i': i},dims=['k','tile','j','i'])
[28]:
print(kapredi_DA.dims)
print(kapredi_DA.shape)
('k', 'tile', 'j', 'i')
(50, 13, 90, 90)
Method 2: llc_tiles_to_xda¶
kapredi is a 4D array (depth, tile, j, i) and therefore we need to pass llc_tiles_to_xda a similarly-dimensioned DataArray or tell the subroutine that the 4th dimension is depth
[29]:
# use similarly-dimensioned DataArray: hFacC
kapredi_DA2 = ecco.llc_tiles_to_xda(kapredi, var_type='c',grid_da=ecco_grid.hFacC)
print(kapredi_DA2.dims)
print(kapredi_DA2.coords)
('k', 'tile', 'j', 'i')
Coordinates:
* k (k) int64 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* j (j) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int64 0 1 2 3 4 5 6 7 8 9 10 11 12
XC (tile, j, i) float32 -111.60647 -111.303 ... -105.58465 -111.86579
YC (tile, j, i) float32 -88.24259 -88.382515 ... -88.07871 -88.10267
CS (tile, j, i) float32 0.06157813 0.06675376 ... -0.9983638
SN (tile, j, i) float32 -0.99810225 -0.9977695 ... -0.057182025
Z (k) float32 -5.0 -15.0 -25.0 -35.0 ... -5039.25 -5461.25 -5906.25
Depth (tile, j, i) float32 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
rA (tile, j, i) float32 362256450.0 363300960.0 ... 361119100.0
PHrefC (k) float32 49.05 147.15 245.25 ... 49435.043 53574.863 57940.312
drF (k) float32 10.0 10.0 10.0 10.0 10.0 ... 387.5 410.5 433.5 456.5
hFacC (k, tile, j, i) float32 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
maskC (k, tile, j, i) bool False False False False ... False False False
[30]:
# specify that the 4th dimension should be depth
kapredi_DA3 = ecco.llc_tiles_to_xda(kapredi, var_type='c', dim4='depth')
print(kapredi_DA3.dims)
print(kapredi_DA3.coords)
('k', 'tile', 'j', 'i')
Coordinates:
* k (k) int64 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* tile (tile) int64 0 1 2 3 4 5 6 7 8 9 10 11 12
* j (j) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int64 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
Parting thoughts¶
read_llc_to_tiles can also be used to read ECCO ‘.data’ generated when re-running the ECCO model.
Converting from numpy ndarrays or xarray DataArrays in the ‘tile’ format to a 5-faces format or compact format can be made with routines like: llc_tiles_to_compact and llc_tiles_to_faces
llc_tiles_to_faces¶
[31]:
help(ecco.llc_tiles_to_faces)
Help on function llc_tiles_to_faces in module ecco_v4_py.llc_array_conversion:
llc_tiles_to_faces(data_tiles, less_output=False)
Converts an array of 13 'tiles' from the lat-lon-cap grid
and rearranges them to 5 faces. Faces 1,2,4, and 5 are approximately
lat-lon while face 3 is the Arctic 'cap'
Parameters
----------
data_tiles :
An array of dimension 13 x nl x nk x llc x llc
If dimensions nl or nk are singular, they are not included
as dimensions of data_tiles
less_output : boolean
A debugging flag. False = less debugging output
Default: False
Returns
-------
F : dict
a dictionary containing the five lat-lon-cap faces
F[n] is a numpy array of face n, n in [1..5]
dimensions of each 2D slice of F
- f1,f2: 3*llc x llc
- f3: llc x llc
- f4,f5: llc x 3*llc
llc_tiles_to_compact¶
[32]:
help(ecco.llc_tiles_to_compact)
Help on function llc_tiles_to_compact in module ecco_v4_py.llc_array_conversion:
llc_tiles_to_compact(data_tiles, less_output=False)
Converts a numpy binary array in the 'compact' format of the
lat-lon-cap (LLC) grids and converts it to the '13 tiles' format
of the LLC grids.
Parameters
----------
data_tiles : ndarray
a numpy array organized by, at most,
13 tiles x nl x nk x llc x llc
where dimensions 'nl' and 'nk' are optional.
less_output : boolean, optional, default False
A debugging flag. False = less debugging output
Returns
-------
data_compact : ndarray
a numpy array of dimension nl x nk x 13*llc x llc
Note
----
If dimensions nl or nk are singular, they are not included
as dimensions in data_compact