Loading the ECCOv4 state estimate fields on the native model grid¶
Objectives¶
Introduce several methods for opening and loading the ECCOv4 state estimate files on the native model grid.
Introduction¶
ECCOv4 native-grid state estimate fields are packaged together as NetCDF files. The ECCOv4 release 4 output files are available from PO.DAAC Cloud via NASA Earthdata, see this tutorial for more information on access. If you already have a NASA Earthdata account, you can download and use the ecco_download module to access ECCOv4r4 datasets, specifying the dataset ShortName you want to download.
For this tutorial, you will need to have downloaded the grid file used in the previous tutorial. You will also need to have downloaded the monthly mean SSH, OBP, and temperature/salinity datasets from 2010-2012. The ShortNames for these datasets are ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4, ECCO_L4_OBP_LLC0090GRID_MONTHLY_V4R4, and ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4.
NetCDF File Format¶
As of March 2023, ECCOv4 release 4 state estimate fields are distributed through PO.DAAC/ NASA Earthdata Cloud, and are stored as NetCDF files, with each file containing one granule. Most datasets contain a few related fields, and a granule is a dataset entry at a single time entry: a monthly mean, daily mean, or a snapshot at 0Z on a given day.
Typical file sizes in the LLC90 grid depend on how many fields are contained in a given dataset:
3D fields: 17-62 MB (2-8 fields x 1 time x 50 levels x 13 tiles x 90 j x 90 i)
2D fields: 5-7 MB (1-12 fields x 1 time x 1 level x 13 tiles x 90 j x 90 i)
Notice that there is a not a very large difference in the range of 2D file sizes between a granule of a dataset having 1 field (5 MB) and another dataset having 12 fields (7 MB). This is because most of the space in individual 2D files is occupied by grid parameters. One nice feature of using the xarray and Dask libraries is that we do not have to load the entire file contents into RAM to work with them. The glob and xarray libraries can also be used together to load multiple
files (times) into your workspace as a single dataset, concatenating the data fields without repeating the grid parameters.
Open/view one ECCOv4 NetCDF file¶
In ECCO NetCDF files, all 13 tiles for a given year are aggregated into a single file. Therefore, we can use the open_dataset routine from xarray to open a single NetCDF variable file.
First set up the environment, load model grid parameters.¶
[1]:
import numpy as np
import xarray as xr
import sys
import matplotlib.pyplot as plt
%matplotlib inline
[2]:
## Import the ecco_v4_py library into Python
## =========================================
## -- If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. For example, if your ecco_v4_py
## files are in /Users/ifenty/ECCOv4-py/ecco_v4_py, then use:
from os.path import join,expanduser
user_home_dir = expanduser('~')
sys.path.append(join(user_home_dir,'ECCOv4-py'))
import ecco_v4_py as ecco
[3]:
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## define a high-level directory for ECCO fields
## currently set to ~/Downloads/ECCO_V4r4_PODAAC, the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join(user_home_dir,'Downloads','ECCO_V4r4_PODAAC')
## define the directory with the model grid
grid_shortname = "ECCO_L4_GEOMETRY_LLC0090GRID_V4R4"
grid_dir = join(ECCO_dir,grid_shortname)
## load the grid
grid_file = "GRID_GEOMETRY_ECCO_V4r4_native_llc0090.nc"
grid = xr.open_dataset(join(grid_dir,grid_file))
Open a single ECCOv4 variable NetCDF file using open_dataset¶
[4]:
SSH_monthly_shortname = "ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4"
SSH_dir = join(ECCO_dir,SSH_monthly_shortname)
SSH_file = "SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc"
SSH_dataset = xr.open_dataset(join(SSH_dir,SSH_file))
SSH_dataset
[4]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, tile: 13, time: 1, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00
... ...
YC (tile, j, i) float32 ...
XG (tile, j_g, i_g) float32 ...
YG (tile, j_g, i_g) float32 ...
time_bnds (time, nv) datetime64[ns] 2010-01-01 2010-02-01
XC_bnds (tile, j, i, nb) float32 ...
YC_bnds (tile, j, i, nb) float32 ...
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 ...
SSHIBC (time, tile, j, i) float32 ...
SSHNOIBC (time, tile, j, i) float32 ...
ETAN (time, tile, j, i) float32 ...
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- tile: 13
- time: 1
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000'], dtype='datetime64[ns]')
- XC(tile, j, i)float32...
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
[105300 values with dtype=float32]
- YC(tile, j, i)float32...
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
[105300 values with dtype=float32]
- XG(tile, j_g, i_g)float32...
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
[105300 values with dtype=float32]
- YG(tile, j_g, i_g)float32...
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
[105300 values with dtype=float32]
- time_bnds(time, nv)datetime64[ns]...
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
array([['2010-01-01T00:00:00.000000000', '2010-02-01T00:00:00.000000000']], dtype='datetime64[ns]') - XC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
[421200 values with dtype=float32]
- YC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
[421200 values with dtype=float32]
- SSH(time, tile, j, i)float32...
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
[105300 values with dtype=float32]
- SSHIBC(time, tile, j, i)float32...
- long_name :
- The inverted barometer (IB) correction to sea surface height due to atmospheric pressure loading
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Not an SSH itself, but a correction to model sea level anomaly (ETAN) required to account for the static part of sea surface displacement by atmosphere pressure loading: SSH = SSHNOIBC - SSHIBC. Note: Use SSH for model-data comparisons with altimetry data products that DO apply the IB correction and SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -0.30144819617271423
- valid_max :
- 0.5248842239379883
[105300 values with dtype=float32]
- SSHNOIBC(time, tile, j, i)float32...
- long_name :
- Sea surface height anomaly without the inverted barometer (IB) correction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Sea surface height anomaly above the geoid without the inverse barometer (IB) correction, suitable for comparisons with altimetry sea surface height data products that do NOT apply the inverse barometer (IB) correction. Note: SSHNOIBC is calculated by correcting model sea level anomaly ETAN for two effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). In ECCO Version 4 Release 4 the model is forced with atmospheric pressure loading. SSHNOIBC does not correct for the static part of the effect of atmosphere pressure loading on sea surface height (the so-called inverse barometer (IB) correction). Use SSH for comparisons with altimetry data products that DO apply the IB correction.
- valid_min :
- -1.6654272079467773
- valid_max :
- 1.4550364017486572
[105300 values with dtype=float32]
- ETAN(time, tile, j, i)float32...
- long_name :
- Model sea level anomaly
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Model sea level anomaly WITHOUT corrections for global mean density (steric) changes, inverted barometer effect, or volume displacement due to submerged sea-ice and snow . Note: ETAN should NOT be used for comparisons with altimetry data products because ETAN is NOT corrected for (a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) nor (b) sea level displacement due to submerged sea-ice and snow (see sIceLoad). These corrections ARE made for the variables SSH and SSHNOIBC.
- valid_min :
- -8.304216384887695
- valid_max :
- 1.460192084312439
[105300 values with dtype=float32]
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:27
- date_issued :
- 2020-12-16T18:07:27
- date_metadata_modified :
- 2021-03-15T21:56:12
- date_modified :
- 2021-03-15T21:56:12
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3
This file (granule) contains four data variables, all related to sea surface height but with important differences. Let’s look at the SSH variable. This is the dynamic sea surface height anomaly, analogous to the gridded dynamic topography (MDT+SLA) that you would obtain from gridded satellite altimetry products (e.g., from JPL MEaSUREs, Copernicus (formerly AVISO)).
[5]:
# look at structure/attributes of a single variable
SSH_dataset.SSH
[5]:
<xarray.DataArray 'SSH' (time: 1, tile: 13, j: 90, i: 90)>
[105300 values with dtype=float32]
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00
XC (tile, j, i) float32 ...
YC (tile, j, i) float32 ...
Attributes:
long_name: Dynamic sea surface height anomaly
units: m
coverage_content_type: modelResult
standard_name: sea_surface_height_above_geoid
comment: Dynamic sea surface height anomaly above the geoi...
valid_min: -1.8805772066116333
valid_max: 1.4207719564437866- time: 1
- tile: 13
- j: 90
- i: 90
- ...
[105300 values with dtype=float32]
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000'], dtype='datetime64[ns]')
- XC(tile, j, i)float32...
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
[105300 values with dtype=float32]
- YC(tile, j, i)float32...
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
[105300 values with dtype=float32]
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
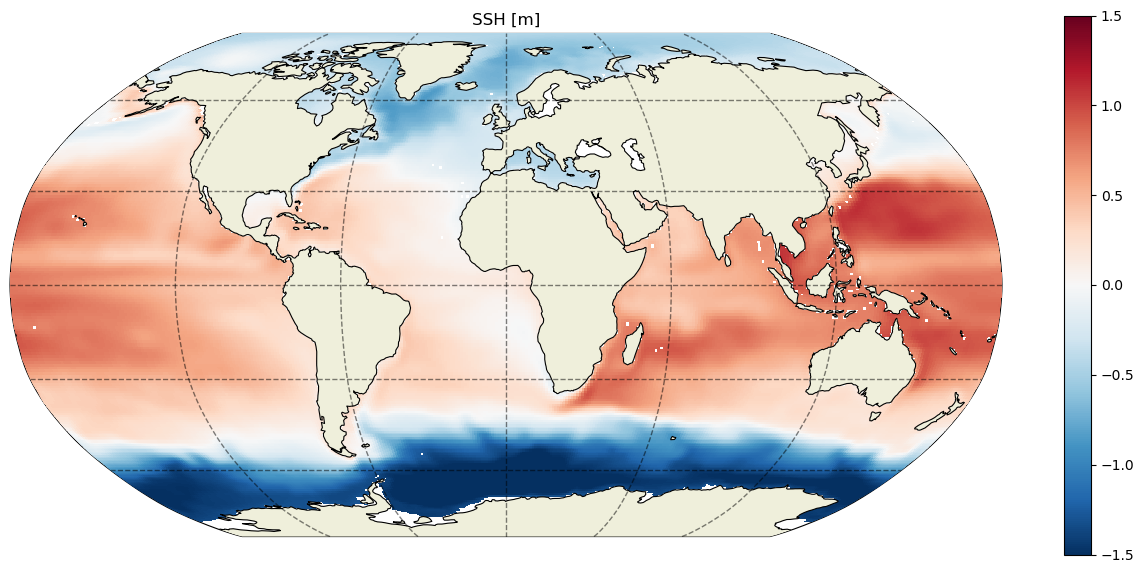
Let’s plot the SSH in this file:
[6]:
SSH = SSH_dataset.SSH
# mask to nan where hFacC(k=0) = 0
SSH = SSH.where(grid.hFacC.isel(k=0))
fig = plt.figure(figsize=(16,7))
ecco.plot_proj_to_latlon_grid(grid.XC, grid.YC, SSH, show_colorbar=True, cmin=-1.5, cmax=1.5);plt.title('SSH [m]');
-179.875 179.875
-180.0 180.0
-89.875 89.875
-90.0 90.0
Alternatively: use glob module to get file name¶
The open_dataset function in xarray requires an exact file name as input. Some of the file names of specific granules are quite long, and looking up the file name each time we want to use a new dataset is a little annoying.
This is where the glob module (generally included in Python distributions) can help us. It takes as input a pattern-matching expression for file names (the same as you would pass to ls in a Unix shell), and returns a Python list of those file names. In this case, we know that we want the Jan 2010 monthly mean SSH, and granule file names for a certain time always contain the expression YYYY-MM (for monthly mean files) or YYYY-MM-DD (for daily mean/snapshot files). So we can get the same
result as above knowing only the dataset ShortName and the month we want to plot.
[7]:
SSH_monthly_shortname = "ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4"
SSH_dir = join(ECCO_dir,SSH_monthly_shortname)
import glob
SSH_files = glob.glob(join(SSH_dir,'*_2010-01_*.nc'))
SSH_dataset = xr.open_dataset(SSH_files[0])
SSH_dataset
[7]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, tile: 13, time: 1, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00
... ...
YC (tile, j, i) float32 ...
XG (tile, j_g, i_g) float32 ...
YG (tile, j_g, i_g) float32 ...
time_bnds (time, nv) datetime64[ns] 2010-01-01 2010-02-01
XC_bnds (tile, j, i, nb) float32 ...
YC_bnds (tile, j, i, nb) float32 ...
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 ...
SSHIBC (time, tile, j, i) float32 ...
SSHNOIBC (time, tile, j, i) float32 ...
ETAN (time, tile, j, i) float32 ...
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- tile: 13
- time: 1
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000'], dtype='datetime64[ns]')
- XC(tile, j, i)float32...
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
[105300 values with dtype=float32]
- YC(tile, j, i)float32...
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
[105300 values with dtype=float32]
- XG(tile, j_g, i_g)float32...
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
[105300 values with dtype=float32]
- YG(tile, j_g, i_g)float32...
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
[105300 values with dtype=float32]
- time_bnds(time, nv)datetime64[ns]...
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
array([['2010-01-01T00:00:00.000000000', '2010-02-01T00:00:00.000000000']], dtype='datetime64[ns]') - XC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
[421200 values with dtype=float32]
- YC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
[421200 values with dtype=float32]
- SSH(time, tile, j, i)float32...
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
[105300 values with dtype=float32]
- SSHIBC(time, tile, j, i)float32...
- long_name :
- The inverted barometer (IB) correction to sea surface height due to atmospheric pressure loading
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Not an SSH itself, but a correction to model sea level anomaly (ETAN) required to account for the static part of sea surface displacement by atmosphere pressure loading: SSH = SSHNOIBC - SSHIBC. Note: Use SSH for model-data comparisons with altimetry data products that DO apply the IB correction and SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -0.30144819617271423
- valid_max :
- 0.5248842239379883
[105300 values with dtype=float32]
- SSHNOIBC(time, tile, j, i)float32...
- long_name :
- Sea surface height anomaly without the inverted barometer (IB) correction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Sea surface height anomaly above the geoid without the inverse barometer (IB) correction, suitable for comparisons with altimetry sea surface height data products that do NOT apply the inverse barometer (IB) correction. Note: SSHNOIBC is calculated by correcting model sea level anomaly ETAN for two effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). In ECCO Version 4 Release 4 the model is forced with atmospheric pressure loading. SSHNOIBC does not correct for the static part of the effect of atmosphere pressure loading on sea surface height (the so-called inverse barometer (IB) correction). Use SSH for comparisons with altimetry data products that DO apply the IB correction.
- valid_min :
- -1.6654272079467773
- valid_max :
- 1.4550364017486572
[105300 values with dtype=float32]
- ETAN(time, tile, j, i)float32...
- long_name :
- Model sea level anomaly
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Model sea level anomaly WITHOUT corrections for global mean density (steric) changes, inverted barometer effect, or volume displacement due to submerged sea-ice and snow . Note: ETAN should NOT be used for comparisons with altimetry data products because ETAN is NOT corrected for (a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) nor (b) sea level displacement due to submerged sea-ice and snow (see sIceLoad). These corrections ARE made for the variables SSH and SSHNOIBC.
- valid_min :
- -8.304216384887695
- valid_max :
- 1.460192084312439
[105300 values with dtype=float32]
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:27
- date_issued :
- 2020-12-16T18:07:27
- date_metadata_modified :
- 2021-03-15T21:56:12
- date_modified :
- 2021-03-15T21:56:12
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3
We can see that the time coordinate is in the correct month.
Opening a subset of a single ECCOv4 variable NetCDF file¶
Once an xarray dataset is opened in the workspace (before it is loaded into actual memory/RAM), it can be subset so that when you actually perform computations, they do not need to involve the entire dataset. This can be done using isel or sel. isel accepts indices, which can be expressed as key/value pairs individually or as a dictionary of key/value pairs. The same is true of sel, but it accepts dimension coordinates instead of indices; most dimensional coordinates in
ECCOv4 output files are given as indices anyway, so there is not much difference (except for subsetting in time).
In the following example we open the monthly mean temperature/salinity file for Jan 2010, then create a subset for tiles 7,8,9 and depth levels 0:34.
[8]:
temp_sal_monthly_shortname = "ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"
temp_sal_dir = join(ECCO_dir,temp_sal_monthly_shortname)
temp_sal_files = glob.glob(join(temp_sal_dir,'*_2010-01_*.nc'))
ds_temp_sal = xr.open_dataset(temp_sal_files[0])
tiles_subset = [7,8,9]
k_subset = np.arange(0,34) # includes k indices 0..33 (not including 34, per Python indexing convention)
ds_subset = ds_temp_sal.isel(tile=tiles_subset,k=k_subset)
ds_subset
[8]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 34, k_u: 50, k_l: 50, k_p1: 51, tile: 3, time: 1, nv: 2, nb: 4)
Coordinates: (12/22)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 ... 24 25 26 27 28 29 30 31 32 33
* k_u (k_u) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 -10.0 -20.0 -30.0 ... -5.678e+03 -6.134e+03
Zl (k_l) float32 0.0 -10.0 -20.0 ... -5.244e+03 -5.678e+03
time_bnds (time, nv) datetime64[ns] 2010-01-01 2010-02-01
XC_bnds (tile, j, i, nb) float32 142.0 142.0 142.4 ... -115.0 -115.3
YC_bnds (tile, j, i, nb) float32 67.5 67.4 67.43 ... -80.47 -80.41 -80.41
Z_bnds (k, nv) float32 0.0 -10.0 -10.0 ... -1.461e+03 -1.573e+03
Dimensions without coordinates: nv, nb
Data variables:
THETA (time, k, tile, j, i) float32 ...
SALT (time, k, tile, j, i) float32 ...
Attributes: (12/62)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Ocean Temperature and Salinity - Mo...
uuid: f4291248-4181-11eb-82cd-0cc47a3f446d- i: 90
- i_g: 90
- j: 90
- j_g: 90
- k: 34
- k_u: 50
- k_l: 50
- k_p1: 51
- tile: 3
- time: 1
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 28 29 30 31 32 33
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33]) - k_u(k_u)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- 0.5
- swap_dim :
- Zu
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the bottom face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the bottom surface of the uppermost tracer grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_l(k_l)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- -0.5
- swap_dim :
- Zl
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the top face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the top surface of the uppermost tracer grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_p1(k_p1)int320 1 2 3 4 5 6 ... 45 46 47 48 49 50
- axis :
- Z
- long_name :
- grid index in z for variables at 'w' locations
- c_grid_axis_shift :
- [-0.5 0.5]
- swap_dim :
- Zp1
- comment :
- Includes top of uppermost model tracer cell (k_p1=0) and bottom of lowermost tracer cell (k_p1=51).
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]) - tile(tile)int327 8 9
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([7, 8, 9])
- time(time)datetime64[ns]2010-01-16T12:00:00
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000'], dtype='datetime64[ns]')
- XC(tile, j, i)float32...
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[ 142.16208 , 142.22801 , ..., 142.5 , 142.5 ], [ 142.55579 , 142.70805 , ..., 143.5 , 143.5 ], ..., [-128.55579 , -128.70805 , ..., -129.5 , -129.5 ], [-128.16208 , -128.22801 , ..., -128.5 , -128.5 ]], [[ 142.5 , 142.5 , ..., 142.5 , 142.5 ], [ 143.5 , 143.5 , ..., 143.5 , 143.5 ], ..., [-129.5 , -129.5 , ..., -129.5 , -129.5 ], [-128.5 , -128.5 , ..., -128.5 , -128.5 ]], [[ 142.5 , 142.5 , ..., 75.1804 , 68.39353 ], [ 143.5 , 143.5 , ..., 76.071556, 68.697 ], ..., [-129.5 , -129.5 , ..., -115.58988 , -115.19479 ], [-128.5 , -128.5 , ..., -115.5476 , -115.18083 ]]], dtype=float32) - YC(tile, j, i)float32...
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[ 67.47211 , 67.33552 , ..., 11.438585, 10.458642], [ 67.53387 , 67.37607 , ..., 11.438585, 10.458642], ..., [ 67.53387 , 67.37607 , ..., 11.438585, 10.458642], [ 67.47211 , 67.33552 , ..., 11.438585, 10.458642]], [[ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], ..., [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ]], [[-57.271408, -57.79962 , ..., -88.21676 , -88.24259 ], [-57.271408, -57.79962 , ..., -88.354485, -88.382515], ..., [-57.271408, -57.79962 , ..., -80.50038 , -80.50492 ], [-57.271408, -57.79962 , ..., -80.43542 , -80.43992 ]]], dtype=float32) - XG(tile, j_g, i_g)float32...
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[ 142. , 142. , ..., 142. , 142. ], [ 142.26555 , 142.41107 , ..., 143. , 143. ], ..., [-128.67538 , -128.88104 , ..., -130. , -130. ], [-128.26555 , -128.41107 , ..., -129. , -129. ]], [[ 142. , 142. , ..., 142. , 142. ], [ 143. , 143. , ..., 143. , 143. ], ..., [-130. , -130. , ..., -130. , -130. ], [-129. , -129. , ..., -129. , -129. ]], [[ 142. , 142. , ..., 78.0046 , 71.52787 ], [ 143. , 143. , ..., 79.08203 , 72.084755], ..., [-130. , -130. , ..., -115.81853 , -115.40557 ], [-129. , -129. , ..., -115.76165 , -115.37739 ]]], dtype=float32) - YG(tile, j_g, i_g)float32...
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[ 67.5 , 67.40169 , ..., 11.928692, 10.948437], [ 67.560646, 67.42869 , ..., 11.928692, 10.948437], ..., [ 67.6532 , 67.49435 , ..., 11.928692, 10.948437], [ 67.560646, 67.42869 , ..., 11.928692, 10.948437]], [[ 9.96973 , 8.997536, ..., -55.931786, -56.471046], [ 9.96973 , 8.997536, ..., -55.931786, -56.471046], ..., [ 9.96973 , 8.997536, ..., -55.931786, -56.471046], [ 9.96973 , 8.997536, ..., -55.931786, -56.471046]], [[-57.005695, -57.53605 , ..., -88.12586 , -88.16335 ], [-57.005695, -57.53605 , ..., -88.26202 , -88.30251 ], ..., [-57.005695, -57.53605 , ..., -80.530266, -80.5372 ], [-57.005695, -57.53605 , ..., -80.46337 , -80.470245]]], dtype=float32) - Z(k)float32...
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ -5. , -15. , -25. , -35. , -45. , -55. , -65. , -75.005, -85.025, -95.095, -105.31 , -115.87 , -127.15 , -139.74 , -154.47 , -172.4 , -194.735, -222.71 , -257.47 , -299.93 , -350.68 , -409.93 , -477.47 , -552.71 , -634.735, -722.4 , -814.47 , -909.74 , -1007.155, -1105.905, -1205.535, -1306.205, -1409.15 , -1517.095], dtype=float32) - Zp1(k_p1)float32...
- long_name :
- depth of tracer grid cell interface
- units :
- m
- positive :
- up
- comment :
- Contains one element more than the number of vertical layers. First element is 0m, the depth of the upper interface of the surface grid cell. Last element is the depth of the lower interface of the deepest grid cell.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zu(k_u)float32...
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the bottom face of tracer grid cells
- comment :
- First element is -10m, the depth of the bottom face of the first tracer grid cell. Last element is the depth of the bottom face of the deepest grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
array([ -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zl(k_l)float32...
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the top face of tracer grid cells
- comment :
- First element is 0m, the depth of the top face of the first tracer grid cell (ocean surface). Last element is the depth of the top face of the deepest grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. ], dtype=float32) - time_bnds(time, nv)datetime64[ns]...
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
array([['2010-01-01T00:00:00.000000000', '2010-02-01T00:00:00.000000000']], dtype='datetime64[ns]') - XC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
array([[[[ 142. , ..., 142.26555 ], ..., [ 142. , ..., 143. ]], ..., [[-128.26555 , ..., -128. ], ..., [-129. , ..., -128. ]]], ..., [[[ 142. , ..., 143. ], ..., [ 71.52787 , ..., 72.084755]], ..., [[-129. , ..., -128. ], ..., [-115.37739 , ..., -115.34945 ]]]], dtype=float32) - YC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
array([[[[ 67.5 , ..., 67.560646], ..., [ 10.948437, ..., 10.948437]], ..., [[ 67.560646, ..., 67.5 ], ..., [ 10.948437, ..., 10.948437]]], ..., [[[-57.005695, ..., -57.005695], ..., [-88.16335 , ..., -88.30251 ]], ..., [[-57.005695, ..., -57.005695], ..., [-80.470245, ..., -80.40726 ]]]], dtype=float32) - Z_bnds(k, nv)float32...
- comment :
- One pair of depths for each vertical level.
- coverage_content_type :
- coordinate
- long_name :
- depths of tracer grid cell upper and lower interfaces
array([[ 0. , -10. ], [ -10. , -20. ], [ -20. , -30. ], [ -30. , -40. ], [ -40. , -50. ], [ -50. , -60. ], [ -60. , -70. ], [ -70. , -80.01 ], [ -80.01 , -90.04 ], [ -90.04 , -100.15 ], [ -100.15 , -110.47 ], [ -110.47 , -121.270004], [ -121.270004, -133.03 ], [ -133.03 , -146.45 ], [ -146.45 , -162.48999 ], [ -162.48999 , -182.31 ], [ -182.31 , -207.16 ], [ -207.16 , -238.26001 ], [ -238.26001 , -276.68 ], [ -276.68 , -323.18 ], [ -323.18 , -378.18 ], [ -378.18 , -441.68 ], [ -441.68 , -513.26 ], [ -513.26 , -592.16003 ], [ -592.16003 , -677.31006 ], [ -677.31006 , -767.49005 ], [ -767.49005 , -861.4501 ], [ -861.4501 , -958.0301 ], [ -958.0301 , -1056.28 ], [-1056.28 , -1155.53 ], [-1155.53 , -1255.54 ], [-1255.54 , -1356.87 ], [-1356.87 , -1461.4299 ], [-1461.4299 , -1572.7599 ]], dtype=float32)
- THETA(time, k, tile, j, i)float32...
- long_name :
- Potential temperature
- units :
- degree_C
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_potential_temperature
- comment :
- Sea water potential temperature is the temperature a parcel of sea water would have if moved adiabatically to sea level pressure. Note: the equation of state is a modified UNESCO formula by Jackett and McDougall (1995), which uses the model variable potential temperature as input assuming a horizontally and temporally constant pressure of $p_0=-g \rho_{0} z$.
- valid_min :
- -2.2909388542175293
- valid_max :
- 36.032955169677734
[826200 values with dtype=float32]
- SALT(time, k, tile, j, i)float32...
- long_name :
- Salinity
- units :
- 1e-3
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_salinity
- valid_min :
- 17.106637954711914
- valid_max :
- 41.26802444458008
- comment :
- Defined using CF convention 'Sea water salinity is the salt content of sea water, often on the Practical Salinity Scale of 1978. However, the unqualified term 'salinity' is generic and does not necessarily imply any particular method of calculation. The units of salinity are dimensionless and the units attribute should normally be given as 1e-3 or 0.001 i.e. parts per thousand.' see https://cfconventions.org/Data/cf-standard-names/73/build/cf-standard-name-table.html
[826200 values with dtype=float32]
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-18T14:39:56
- date_issued :
- 2020-12-18T14:39:56
- date_metadata_modified :
- 2021-03-15T21:55:41
- date_modified :
- 2021-03-15T21:55:41
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- geospatial_vertical_max :
- 0.0
- geospatial_vertical_min :
- -6134.5
- geospatial_vertical_positive :
- up
- geospatial_vertical_resolution :
- variable
- geospatial_vertical_units :
- meter
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-OTS44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > OCEAN TEMPERATURE > POTENTIAL TEMPERATURE, EARTH SCIENCE > OCEANS > SALINITY/DENSITY > SALINITY, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- OCEAN_TEMPERATURE_SALINITY_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged ocean potential temperature and salinity on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Ocean Temperature and Salinity - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- f4291248-4181-11eb-82cd-0cc47a3f446d
As expected, ds_subset has 3 tiles and 34 vertical levels. We can do the same thing, but pass both tile and k subset indices in a single dictionary; this is more convenient if we use these subsetting indices multiple times.
[9]:
tiles_subset = [7,8,9]
k_subset = np.arange(0,34) # includes k indices 0..33 (not including 34, per Python indexing convention)
dict_subset_ind = {'tile':tiles_subset,'k':k_subset}
ds_subset = ds_temp_sal.isel(dict_subset_ind)
ds_subset
[9]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 34, k_u: 50, k_l: 50, k_p1: 51, tile: 3, time: 1, nv: 2, nb: 4)
Coordinates: (12/22)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 ... 24 25 26 27 28 29 30 31 32 33
* k_u (k_u) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 -10.0 -20.0 -30.0 ... -5.678e+03 -6.134e+03
Zl (k_l) float32 0.0 -10.0 -20.0 ... -5.244e+03 -5.678e+03
time_bnds (time, nv) datetime64[ns] 2010-01-01 2010-02-01
XC_bnds (tile, j, i, nb) float32 142.0 142.0 142.4 ... -115.0 -115.3
YC_bnds (tile, j, i, nb) float32 67.5 67.4 67.43 ... -80.47 -80.41 -80.41
Z_bnds (k, nv) float32 0.0 -10.0 -10.0 ... -1.461e+03 -1.573e+03
Dimensions without coordinates: nv, nb
Data variables:
THETA (time, k, tile, j, i) float32 ...
SALT (time, k, tile, j, i) float32 ...
Attributes: (12/62)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Ocean Temperature and Salinity - Mo...
uuid: f4291248-4181-11eb-82cd-0cc47a3f446d- i: 90
- i_g: 90
- j: 90
- j_g: 90
- k: 34
- k_u: 50
- k_l: 50
- k_p1: 51
- tile: 3
- time: 1
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 28 29 30 31 32 33
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33]) - k_u(k_u)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- 0.5
- swap_dim :
- Zu
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the bottom face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the bottom surface of the uppermost tracer grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_l(k_l)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- -0.5
- swap_dim :
- Zl
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the top face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the top surface of the uppermost tracer grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_p1(k_p1)int320 1 2 3 4 5 6 ... 45 46 47 48 49 50
- axis :
- Z
- long_name :
- grid index in z for variables at 'w' locations
- c_grid_axis_shift :
- [-0.5 0.5]
- swap_dim :
- Zp1
- comment :
- Includes top of uppermost model tracer cell (k_p1=0) and bottom of lowermost tracer cell (k_p1=51).
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]) - tile(tile)int327 8 9
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([7, 8, 9])
- time(time)datetime64[ns]2010-01-16T12:00:00
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000'], dtype='datetime64[ns]')
- XC(tile, j, i)float32...
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[ 142.16208 , 142.22801 , ..., 142.5 , 142.5 ], [ 142.55579 , 142.70805 , ..., 143.5 , 143.5 ], ..., [-128.55579 , -128.70805 , ..., -129.5 , -129.5 ], [-128.16208 , -128.22801 , ..., -128.5 , -128.5 ]], [[ 142.5 , 142.5 , ..., 142.5 , 142.5 ], [ 143.5 , 143.5 , ..., 143.5 , 143.5 ], ..., [-129.5 , -129.5 , ..., -129.5 , -129.5 ], [-128.5 , -128.5 , ..., -128.5 , -128.5 ]], [[ 142.5 , 142.5 , ..., 75.1804 , 68.39353 ], [ 143.5 , 143.5 , ..., 76.071556, 68.697 ], ..., [-129.5 , -129.5 , ..., -115.58988 , -115.19479 ], [-128.5 , -128.5 , ..., -115.5476 , -115.18083 ]]], dtype=float32) - YC(tile, j, i)float32...
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[ 67.47211 , 67.33552 , ..., 11.438585, 10.458642], [ 67.53387 , 67.37607 , ..., 11.438585, 10.458642], ..., [ 67.53387 , 67.37607 , ..., 11.438585, 10.458642], [ 67.47211 , 67.33552 , ..., 11.438585, 10.458642]], [[ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], ..., [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ], [ 9.482398, 8.516253, ..., -56.2021 , -56.73891 ]], [[-57.271408, -57.79962 , ..., -88.21676 , -88.24259 ], [-57.271408, -57.79962 , ..., -88.354485, -88.382515], ..., [-57.271408, -57.79962 , ..., -80.50038 , -80.50492 ], [-57.271408, -57.79962 , ..., -80.43542 , -80.43992 ]]], dtype=float32) - XG(tile, j_g, i_g)float32...
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[ 142. , 142. , ..., 142. , 142. ], [ 142.26555 , 142.41107 , ..., 143. , 143. ], ..., [-128.67538 , -128.88104 , ..., -130. , -130. ], [-128.26555 , -128.41107 , ..., -129. , -129. ]], [[ 142. , 142. , ..., 142. , 142. ], [ 143. , 143. , ..., 143. , 143. ], ..., [-130. , -130. , ..., -130. , -130. ], [-129. , -129. , ..., -129. , -129. ]], [[ 142. , 142. , ..., 78.0046 , 71.52787 ], [ 143. , 143. , ..., 79.08203 , 72.084755], ..., [-130. , -130. , ..., -115.81853 , -115.40557 ], [-129. , -129. , ..., -115.76165 , -115.37739 ]]], dtype=float32) - YG(tile, j_g, i_g)float32...
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[ 67.5 , 67.40169 , ..., 11.928692, 10.948437], [ 67.560646, 67.42869 , ..., 11.928692, 10.948437], ..., [ 67.6532 , 67.49435 , ..., 11.928692, 10.948437], [ 67.560646, 67.42869 , ..., 11.928692, 10.948437]], [[ 9.96973 , 8.997536, ..., -55.931786, -56.471046], [ 9.96973 , 8.997536, ..., -55.931786, -56.471046], ..., [ 9.96973 , 8.997536, ..., -55.931786, -56.471046], [ 9.96973 , 8.997536, ..., -55.931786, -56.471046]], [[-57.005695, -57.53605 , ..., -88.12586 , -88.16335 ], [-57.005695, -57.53605 , ..., -88.26202 , -88.30251 ], ..., [-57.005695, -57.53605 , ..., -80.530266, -80.5372 ], [-57.005695, -57.53605 , ..., -80.46337 , -80.470245]]], dtype=float32) - Z(k)float32...
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ -5. , -15. , -25. , -35. , -45. , -55. , -65. , -75.005, -85.025, -95.095, -105.31 , -115.87 , -127.15 , -139.74 , -154.47 , -172.4 , -194.735, -222.71 , -257.47 , -299.93 , -350.68 , -409.93 , -477.47 , -552.71 , -634.735, -722.4 , -814.47 , -909.74 , -1007.155, -1105.905, -1205.535, -1306.205, -1409.15 , -1517.095], dtype=float32) - Zp1(k_p1)float320.0 -10.0 ... -5.678e+03 -6.134e+03
- long_name :
- depth of tracer grid cell interface
- units :
- m
- positive :
- up
- comment :
- Contains one element more than the number of vertical layers. First element is 0m, the depth of the upper interface of the surface grid cell. Last element is the depth of the lower interface of the deepest grid cell.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zu(k_u)float32-10.0 -20.0 ... -6.134e+03
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the bottom face of tracer grid cells
- comment :
- First element is -10m, the depth of the bottom face of the first tracer grid cell. Last element is the depth of the bottom face of the deepest grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
array([ -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zl(k_l)float320.0 -10.0 ... -5.244e+03 -5.678e+03
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the top face of tracer grid cells
- comment :
- First element is 0m, the depth of the top face of the first tracer grid cell (ocean surface). Last element is the depth of the top face of the deepest grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. ], dtype=float32) - time_bnds(time, nv)datetime64[ns]2010-01-01 2010-02-01
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
array([['2010-01-01T00:00:00.000000000', '2010-02-01T00:00:00.000000000']], dtype='datetime64[ns]') - XC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
array([[[[ 142. , ..., 142.26555 ], ..., [ 142. , ..., 143. ]], ..., [[-128.26555 , ..., -128. ], ..., [-129. , ..., -128. ]]], ..., [[[ 142. , ..., 143. ], ..., [ 71.52787 , ..., 72.084755]], ..., [[-129. , ..., -128. ], ..., [-115.37739 , ..., -115.34945 ]]]], dtype=float32) - YC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
array([[[[ 67.5 , ..., 67.560646], ..., [ 10.948437, ..., 10.948437]], ..., [[ 67.560646, ..., 67.5 ], ..., [ 10.948437, ..., 10.948437]]], ..., [[[-57.005695, ..., -57.005695], ..., [-88.16335 , ..., -88.30251 ]], ..., [[-57.005695, ..., -57.005695], ..., [-80.470245, ..., -80.40726 ]]]], dtype=float32) - Z_bnds(k, nv)float32...
- comment :
- One pair of depths for each vertical level.
- coverage_content_type :
- coordinate
- long_name :
- depths of tracer grid cell upper and lower interfaces
array([[ 0. , -10. ], [ -10. , -20. ], [ -20. , -30. ], [ -30. , -40. ], [ -40. , -50. ], [ -50. , -60. ], [ -60. , -70. ], [ -70. , -80.01 ], [ -80.01 , -90.04 ], [ -90.04 , -100.15 ], [ -100.15 , -110.47 ], [ -110.47 , -121.270004], [ -121.270004, -133.03 ], [ -133.03 , -146.45 ], [ -146.45 , -162.48999 ], [ -162.48999 , -182.31 ], [ -182.31 , -207.16 ], [ -207.16 , -238.26001 ], [ -238.26001 , -276.68 ], [ -276.68 , -323.18 ], [ -323.18 , -378.18 ], [ -378.18 , -441.68 ], [ -441.68 , -513.26 ], [ -513.26 , -592.16003 ], [ -592.16003 , -677.31006 ], [ -677.31006 , -767.49005 ], [ -767.49005 , -861.4501 ], [ -861.4501 , -958.0301 ], [ -958.0301 , -1056.28 ], [-1056.28 , -1155.53 ], [-1155.53 , -1255.54 ], [-1255.54 , -1356.87 ], [-1356.87 , -1461.4299 ], [-1461.4299 , -1572.7599 ]], dtype=float32)
- THETA(time, k, tile, j, i)float32...
- long_name :
- Potential temperature
- units :
- degree_C
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_potential_temperature
- comment :
- Sea water potential temperature is the temperature a parcel of sea water would have if moved adiabatically to sea level pressure. Note: the equation of state is a modified UNESCO formula by Jackett and McDougall (1995), which uses the model variable potential temperature as input assuming a horizontally and temporally constant pressure of $p_0=-g \rho_{0} z$.
- valid_min :
- -2.2909388542175293
- valid_max :
- 36.032955169677734
[826200 values with dtype=float32]
- SALT(time, k, tile, j, i)float32...
- long_name :
- Salinity
- units :
- 1e-3
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_salinity
- valid_min :
- 17.106637954711914
- valid_max :
- 41.26802444458008
- comment :
- Defined using CF convention 'Sea water salinity is the salt content of sea water, often on the Practical Salinity Scale of 1978. However, the unqualified term 'salinity' is generic and does not necessarily imply any particular method of calculation. The units of salinity are dimensionless and the units attribute should normally be given as 1e-3 or 0.001 i.e. parts per thousand.' see https://cfconventions.org/Data/cf-standard-names/73/build/cf-standard-name-table.html
[826200 values with dtype=float32]
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-18T14:39:56
- date_issued :
- 2020-12-18T14:39:56
- date_metadata_modified :
- 2021-03-15T21:55:41
- date_modified :
- 2021-03-15T21:55:41
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- geospatial_vertical_max :
- 0.0
- geospatial_vertical_min :
- -6134.5
- geospatial_vertical_positive :
- up
- geospatial_vertical_resolution :
- variable
- geospatial_vertical_units :
- meter
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-OTS44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > OCEAN TEMPERATURE > POTENTIAL TEMPERATURE, EARTH SCIENCE > OCEANS > SALINITY/DENSITY > SALINITY, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- OCEAN_TEMPERATURE_SALINITY_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged ocean potential temperature and salinity on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Ocean Temperature and Salinity - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- f4291248-4181-11eb-82cd-0cc47a3f446d
Opening multiple files of a single ECCOv4 variable using open_mfdataset¶
We’ve seen that open_dataset opens a single netCDF file in the workspace as an xarray dataset. The open_mfdataset does the same thing, but automatically concatenates multiple netCDF files based on the dimension and coordinate information in each, a very convenient feature!
Let’s open all the monthly mean SSH files for 2010-2011 as a single dataset. Another convenience of open_mfdataset is that it parses pattern-matching strings in the same way that glob does. This allows ECCOv4 files across a range of times to be concatenated and opened in a single command.
[10]:
# two years: 2010 and 2011
SSH_2010_2011 = xr.open_mfdataset(join(SSH_dir,'*_201[0-1]-??_*.nc'))
SSH_2010_2011
[10]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, tile: 13, time: 24, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2011-12-16T12:00:00
... ...
YC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
XG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHNOIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ETAN (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- tile: 13
- time: 24
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2011-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000', '2011-01-16T12:00:00.000000000', '2011-02-15T00:00:00.000000000', '2011-03-16T12:00:00.000000000', '2011-04-16T00:00:00.000000000', '2011-05-16T12:00:00.000000000', '2011-06-16T00:00:00.000000000', '2011-07-16T12:00:00.000000000', '2011-08-16T12:00:00.000000000', '2011-09-16T00:00:00.000000000', '2011-10-16T12:00:00.000000000', '2011-11-16T00:00:00.000000000', '2011-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - XC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray - YC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray - XG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray - YG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray - time_bnds(time, nv)datetime64[ns]dask.array<chunksize=(1, 2), meta=np.ndarray>
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
Array Chunk Bytes 384 B 16 B Shape (24, 2) (1, 2) Count 72 Tasks 24 Chunks Type datetime64[ns] numpy.ndarray - XC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray - YC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 115 Tasks 1 Chunks Type float32 numpy.ndarray
- SSH(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
Array Chunk Bytes 9.64 MiB 411.33 kiB Shape (24, 13, 90, 90) (1, 13, 90, 90) Count 72 Tasks 24 Chunks Type float32 numpy.ndarray - SSHIBC(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- The inverted barometer (IB) correction to sea surface height due to atmospheric pressure loading
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Not an SSH itself, but a correction to model sea level anomaly (ETAN) required to account for the static part of sea surface displacement by atmosphere pressure loading: SSH = SSHNOIBC - SSHIBC. Note: Use SSH for model-data comparisons with altimetry data products that DO apply the IB correction and SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -0.30144819617271423
- valid_max :
- 0.5248842239379883
Array Chunk Bytes 9.64 MiB 411.33 kiB Shape (24, 13, 90, 90) (1, 13, 90, 90) Count 72 Tasks 24 Chunks Type float32 numpy.ndarray - SSHNOIBC(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Sea surface height anomaly without the inverted barometer (IB) correction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Sea surface height anomaly above the geoid without the inverse barometer (IB) correction, suitable for comparisons with altimetry sea surface height data products that do NOT apply the inverse barometer (IB) correction. Note: SSHNOIBC is calculated by correcting model sea level anomaly ETAN for two effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). In ECCO Version 4 Release 4 the model is forced with atmospheric pressure loading. SSHNOIBC does not correct for the static part of the effect of atmosphere pressure loading on sea surface height (the so-called inverse barometer (IB) correction). Use SSH for comparisons with altimetry data products that DO apply the IB correction.
- valid_min :
- -1.6654272079467773
- valid_max :
- 1.4550364017486572
Array Chunk Bytes 9.64 MiB 411.33 kiB Shape (24, 13, 90, 90) (1, 13, 90, 90) Count 72 Tasks 24 Chunks Type float32 numpy.ndarray - ETAN(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Model sea level anomaly
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Model sea level anomaly WITHOUT corrections for global mean density (steric) changes, inverted barometer effect, or volume displacement due to submerged sea-ice and snow . Note: ETAN should NOT be used for comparisons with altimetry data products because ETAN is NOT corrected for (a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) nor (b) sea level displacement due to submerged sea-ice and snow (see sIceLoad). These corrections ARE made for the variables SSH and SSHNOIBC.
- valid_min :
- -8.304216384887695
- valid_max :
- 1.460192084312439
Array Chunk Bytes 9.64 MiB 411.33 kiB Shape (24, 13, 90, 90) (1, 13, 90, 90) Count 72 Tasks 24 Chunks Type float32 numpy.ndarray
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:27
- date_issued :
- 2020-12-16T18:07:27
- date_metadata_modified :
- 2021-03-15T21:56:12
- date_modified :
- 2021-03-15T21:56:12
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3
Notice that SSH_2010_2011 has 24 time records. You can check that the times were concatenated in order:
[11]:
SSH_2010_2011.time
[11]:
<xarray.DataArray 'time' (time: 24)>
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000',
'2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000',
'2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000',
'2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000',
'2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000',
'2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000',
'2011-01-16T12:00:00.000000000', '2011-02-15T00:00:00.000000000',
'2011-03-16T12:00:00.000000000', '2011-04-16T00:00:00.000000000',
'2011-05-16T12:00:00.000000000', '2011-06-16T00:00:00.000000000',
'2011-07-16T12:00:00.000000000', '2011-08-16T12:00:00.000000000',
'2011-09-16T00:00:00.000000000', '2011-10-16T12:00:00.000000000',
'2011-11-16T00:00:00.000000000', '2011-12-16T12:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2011-12-16T12:00:00
Attributes:
long_name: center time of averaging period
axis: T
bounds: time_bnds
coverage_content_type: coordinate
standard_name: time- time: 24
- 2010-01-16T12:00:00 2010-02-15 ... 2011-11-16 2011-12-16T12:00:00
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000', '2011-01-16T12:00:00.000000000', '2011-02-15T00:00:00.000000000', '2011-03-16T12:00:00.000000000', '2011-04-16T00:00:00.000000000', '2011-05-16T12:00:00.000000000', '2011-06-16T00:00:00.000000000', '2011-07-16T12:00:00.000000000', '2011-08-16T12:00:00.000000000', '2011-09-16T00:00:00.000000000', '2011-10-16T12:00:00.000000000', '2011-11-16T00:00:00.000000000', '2011-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - time(time)datetime64[ns]2010-01-16T12:00:00 ... 2011-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000', '2011-01-16T12:00:00.000000000', '2011-02-15T00:00:00.000000000', '2011-03-16T12:00:00.000000000', '2011-04-16T00:00:00.000000000', '2011-05-16T12:00:00.000000000', '2011-06-16T00:00:00.000000000', '2011-07-16T12:00:00.000000000', '2011-08-16T12:00:00.000000000', '2011-09-16T00:00:00.000000000', '2011-10-16T12:00:00.000000000', '2011-11-16T00:00:00.000000000', '2011-12-16T12:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
Notice also that open_mfdataset opens many variables in the form of a dask.array with the content of the variable divided into chunks. You can also use open_mfdataset to open a single netCDF file; however, in small datasets Dask arrays can slow down computations, so this is not always recommended. We will see some of the benefits of opening variables as Dask arrays soon.
We can also open the monthly mean SSH for all available times in the same way.
[12]:
# all years
SSH_all = xr.open_mfdataset(join(SSH_dir,'*.nc'))
SSH_all
[12]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, tile: 13, time: 312, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 1992-01-16T18:00:00 ... 2017-12-16T06:00:00
... ...
YC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
XG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHNOIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ETAN (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 1992-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 1992-01-01T12:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9302811e-400c-11eb-b69e-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- tile: 13
- time: 312
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]1992-01-16T18:00:00 ... 2017-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['1992-01-16T18:00:00.000000000', '1992-02-15T12:00:00.000000000', '1992-03-16T12:00:00.000000000', ..., '2017-10-16T12:00:00.000000000', '2017-11-16T00:00:00.000000000', '2017-12-16T06:00:00.000000000'], dtype='datetime64[ns]') - XC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray - YC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray - XG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray - YG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray - time_bnds(time, nv)datetime64[ns]dask.array<chunksize=(1, 2), meta=np.ndarray>
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
Array Chunk Bytes 4.88 kiB 16 B Shape (312, 2) (1, 2) Count 936 Tasks 312 Chunks Type datetime64[ns] numpy.ndarray - XC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray - YC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 1555 Tasks 1 Chunks Type float32 numpy.ndarray
- SSH(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
Array Chunk Bytes 125.33 MiB 411.33 kiB Shape (312, 13, 90, 90) (1, 13, 90, 90) Count 936 Tasks 312 Chunks Type float32 numpy.ndarray - SSHIBC(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- The inverted barometer (IB) correction to sea surface height due to atmospheric pressure loading
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Not an SSH itself, but a correction to model sea level anomaly (ETAN) required to account for the static part of sea surface displacement by atmosphere pressure loading: SSH = SSHNOIBC - SSHIBC. Note: Use SSH for model-data comparisons with altimetry data products that DO apply the IB correction and SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -0.30144819617271423
- valid_max :
- 0.5248842239379883
Array Chunk Bytes 125.33 MiB 411.33 kiB Shape (312, 13, 90, 90) (1, 13, 90, 90) Count 936 Tasks 312 Chunks Type float32 numpy.ndarray - SSHNOIBC(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Sea surface height anomaly without the inverted barometer (IB) correction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Sea surface height anomaly above the geoid without the inverse barometer (IB) correction, suitable for comparisons with altimetry sea surface height data products that do NOT apply the inverse barometer (IB) correction. Note: SSHNOIBC is calculated by correcting model sea level anomaly ETAN for two effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). In ECCO Version 4 Release 4 the model is forced with atmospheric pressure loading. SSHNOIBC does not correct for the static part of the effect of atmosphere pressure loading on sea surface height (the so-called inverse barometer (IB) correction). Use SSH for comparisons with altimetry data products that DO apply the IB correction.
- valid_min :
- -1.6654272079467773
- valid_max :
- 1.4550364017486572
Array Chunk Bytes 125.33 MiB 411.33 kiB Shape (312, 13, 90, 90) (1, 13, 90, 90) Count 936 Tasks 312 Chunks Type float32 numpy.ndarray - ETAN(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Model sea level anomaly
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Model sea level anomaly WITHOUT corrections for global mean density (steric) changes, inverted barometer effect, or volume displacement due to submerged sea-ice and snow . Note: ETAN should NOT be used for comparisons with altimetry data products because ETAN is NOT corrected for (a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) nor (b) sea level displacement due to submerged sea-ice and snow (see sIceLoad). These corrections ARE made for the variables SSH and SSHNOIBC.
- valid_min :
- -8.304216384887695
- valid_max :
- 1.460192084312439
Array Chunk Bytes 125.33 MiB 411.33 kiB Shape (312, 13, 90, 90) (1, 13, 90, 90) Count 936 Tasks 312 Chunks Type float32 numpy.ndarray
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:10
- date_issued :
- 2020-12-16T18:07:10
- date_metadata_modified :
- 2021-03-15T21:53:43
- date_modified :
- 2021-03-15T21:53:43
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_1992-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 1992-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 1992-01-01T12:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9302811e-400c-11eb-b69e-0cc47a3f49c3
Now we have all 312 monthly mean records of SSH.
Combining datasets using xarray.merge¶
xarray datasets can be merged to allow the user to more easily work with variables that were not contained in the same dataset or file to begin with.
Say we want to look at the monthly mean sea surface height SSH and potential temperature THETA during 2010-2012. We can open the datasets containing them separately, and then merge them.
If we’re not interested in including a certain variable (e.g., salinity SALT, which is a large full depth variable that we don’t need), then we can also avoid including it in the dataset to begin with by passing the drop_variables option to open_dataset or open_mfdataset.
Opening a dataset of SSH and THETA for 2010-2012¶
[13]:
SSH_2010_2012 = xr.open_mfdataset(join(SSH_dir,'*_201[0-2]-??_*.nc'),drop_variables=['SSHIBC','SSHNOIBC','ETAN'])
THETA_2010_2012 = xr.open_mfdataset(join(temp_sal_dir,'*_201[0-2]-??_*.nc'),drop_variables='SALT')
SSH_THETA_2010_2012 = xr.merge([SSH_2010_2012,THETA_2010_2012])
SSH_THETA_2010_2012
[13]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, tile: 13, time: 36, nv: 2, nb: 4, k: 50, k_u: 50, k_l: 50, k_p1: 51)
Coordinates: (12/22)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2012-12-16T12:00:00
... ...
* k_p1 (k_p1) int32 0 1 2 3 4 5 6 7 8 9 ... 42 43 44 45 46 47 48 49 50
Z (k) float32 dask.array<chunksize=(50,), meta=np.ndarray>
Zp1 (k_p1) float32 dask.array<chunksize=(51,), meta=np.ndarray>
Zu (k_u) float32 dask.array<chunksize=(50,), meta=np.ndarray>
Zl (k_l) float32 dask.array<chunksize=(50,), meta=np.ndarray>
Z_bnds (k, nv) float32 dask.array<chunksize=(50, 2), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
THETA (time, k, tile, j, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- tile: 13
- time: 36
- nv: 2
- nb: 4
- k: 50
- k_u: 50
- k_l: 50
- k_p1: 51
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2012-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000', '2011-01-16T12:00:00.000000000', '2011-02-15T00:00:00.000000000', '2011-03-16T12:00:00.000000000', '2011-04-16T00:00:00.000000000', '2011-05-16T12:00:00.000000000', '2011-06-16T00:00:00.000000000', '2011-07-16T12:00:00.000000000', '2011-08-16T12:00:00.000000000', '2011-09-16T00:00:00.000000000', '2011-10-16T12:00:00.000000000', '2011-11-16T00:00:00.000000000', '2011-12-16T12:00:00.000000000', '2012-01-16T12:00:00.000000000', '2012-02-15T12:00:00.000000000', '2012-03-16T12:00:00.000000000', '2012-04-16T00:00:00.000000000', '2012-05-16T12:00:00.000000000', '2012-06-16T00:00:00.000000000', '2012-07-16T12:00:00.000000000', '2012-08-16T12:00:00.000000000', '2012-09-16T00:00:00.000000000', '2012-10-16T12:00:00.000000000', '2012-11-16T00:00:00.000000000', '2012-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - XC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - YC(tile, j, i)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - XG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - YG(tile, j_g, i_g)float32dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 411.33 kiB 411.33 kiB Shape (13, 90, 90) (13, 90, 90) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - time_bnds(time, nv)datetime64[ns]dask.array<chunksize=(1, 2), meta=np.ndarray>
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
Array Chunk Bytes 576 B 16 B Shape (36, 2) (1, 2) Count 290 Tasks 36 Chunks Type datetime64[ns] numpy.ndarray - XC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - YC_bnds(tile, j, i, nb)float32dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
Array Chunk Bytes 1.61 MiB 1.61 MiB Shape (13, 90, 90, 4) (13, 90, 90, 4) Count 176 Tasks 1 Chunks Type float32 numpy.ndarray - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_u(k_u)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- 0.5
- swap_dim :
- Zu
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the bottom face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the bottom surface of the uppermost tracer grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_l(k_l)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- c_grid_axis_shift :
- -0.5
- swap_dim :
- Zl
- coverage_content_type :
- coordinate
- long_name :
- grid index in z corresponding to the top face of tracer grid cells ('w' locations)
- comment :
- First index corresponds to the top surface of the uppermost tracer grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid.
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_p1(k_p1)int320 1 2 3 4 5 6 ... 45 46 47 48 49 50
- axis :
- Z
- long_name :
- grid index in z for variables at 'w' locations
- c_grid_axis_shift :
- [-0.5 0.5]
- swap_dim :
- Zp1
- comment :
- Includes top of uppermost model tracer cell (k_p1=0) and bottom of lowermost tracer cell (k_p1=51).
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]) - Z(k)float32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
Array Chunk Bytes 200 B 200 B Shape (50,) (50,) Count 175 Tasks 1 Chunks Type float32 numpy.ndarray - Zp1(k_p1)float32dask.array<chunksize=(51,), meta=np.ndarray>
- long_name :
- depth of tracer grid cell interface
- units :
- m
- positive :
- up
- comment :
- Contains one element more than the number of vertical layers. First element is 0m, the depth of the upper interface of the surface grid cell. Last element is the depth of the lower interface of the deepest grid cell.
- coverage_content_type :
- coordinate
- standard_name :
- depth
Array Chunk Bytes 204 B 204 B Shape (51,) (51,) Count 175 Tasks 1 Chunks Type float32 numpy.ndarray - Zu(k_u)float32dask.array<chunksize=(50,), meta=np.ndarray>
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the bottom face of tracer grid cells
- comment :
- First element is -10m, the depth of the bottom face of the first tracer grid cell. Last element is the depth of the bottom face of the deepest grid cell. The use of 'u' in the variable name follows the MITgcm convention for ocean variables in which the upper (u) face of a tracer grid cell on the logical grid corresponds to the bottom face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
Array Chunk Bytes 200 B 200 B Shape (50,) (50,) Count 175 Tasks 1 Chunks Type float32 numpy.ndarray - Zl(k_l)float32dask.array<chunksize=(50,), meta=np.ndarray>
- units :
- m
- positive :
- up
- coverage_content_type :
- coordinate
- standard_name :
- depth
- long_name :
- depth of the top face of tracer grid cells
- comment :
- First element is 0m, the depth of the top face of the first tracer grid cell (ocean surface). Last element is the depth of the top face of the deepest grid cell. The use of 'l' in the variable name follows the MITgcm convention for ocean variables in which the lower (l) face of a tracer grid cell on the logical grid corresponds to the top face of the grid cell on the physical grid. In other words, the logical vertical grid of MITgcm ocean variables is inverted relative to the physical vertical grid.
Array Chunk Bytes 200 B 200 B Shape (50,) (50,) Count 175 Tasks 1 Chunks Type float32 numpy.ndarray - Z_bnds(k, nv)float32dask.array<chunksize=(50, 2), meta=np.ndarray>
- comment :
- One pair of depths for each vertical level.
- coverage_content_type :
- coordinate
- long_name :
- depths of tracer grid cell upper and lower interfaces
Array Chunk Bytes 400 B 400 B Shape (50, 2) (50, 2) Count 175 Tasks 1 Chunks Type float32 numpy.ndarray
- SSH(time, tile, j, i)float32dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
Array Chunk Bytes 14.46 MiB 411.33 kiB Shape (36, 13, 90, 90) (1, 13, 90, 90) Count 108 Tasks 36 Chunks Type float32 numpy.ndarray - THETA(time, k, tile, j, i)float32dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
- long_name :
- Potential temperature
- units :
- degree_C
- coverage_content_type :
- modelResult
- standard_name :
- sea_water_potential_temperature
- comment :
- Sea water potential temperature is the temperature a parcel of sea water would have if moved adiabatically to sea level pressure. Note: the equation of state is a modified UNESCO formula by Jackett and McDougall (1995), which uses the model variable potential temperature as input assuming a horizontally and temporally constant pressure of $p_0=-g \rho_{0} z$.
- valid_min :
- -2.2909388542175293
- valid_max :
- 36.032955169677734
Array Chunk Bytes 723.04 MiB 20.08 MiB Shape (36, 50, 13, 90, 90) (1, 50, 13, 90, 90) Count 108 Tasks 36 Chunks Type float32 numpy.ndarray
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:27
- date_issued :
- 2020-12-16T18:07:27
- date_metadata_modified :
- 2021-03-15T21:56:12
- date_modified :
- 2021-03-15T21:56:12
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3
We see three years (36 months) of data and 13 tiles and 50 vertical levels.
Using a preprocess function to open a spatial subset of multiple files¶
Previously we had an example where we opened a single file as a dataset and then subsetted certain indices as a separate (smaller) dataset. What if we want a dataset with multiple times and a spatial subset? We could use open_mfdataset and then subset like we did before, or we can subset before the datasets are concatenated using a preprocess function.
Let’s try it both ways, to obtain a dataset that contains SSH and OBP monthly means during 2010 in a single tile (tile=6). For good measure, we’ll use the time module and pympler package to track the time and memory footprint involved using each method. If you don’t have Pympler installed already, you can use conda install pympler, conda install -c conda-forge pympler, or pip install pympler to get it on your system.
[14]:
import time
from pympler import asizeof
# function to quantify memory footprint of a dataset
def memory_footprint(ds):
s = 0
for i in ds.variables.keys():
s += asizeof.asizeof(ds[i])
return s
# Merge first then subset
start_time = time.time()
memory_footp = 0
SSH_monthly_2010 = xr.open_mfdataset(join(ECCO_dir,'*SSH*MONTHLY*/*_2010-??_*.nc'))
memory_footp += memory_footprint(SSH_monthly_2010)
OBP_monthly_2010 = xr.open_mfdataset(join(ECCO_dir,'*OBP*MONTHLY*/*_2010-??_*.nc'))
memory_footp += memory_footprint(OBP_monthly_2010)
SSH_OBP_monthly_2010 = xr.merge([SSH_monthly_2010,OBP_monthly_2010])
memory_footp += memory_footprint(SSH_OBP_monthly_2010)
SSH_OBP_2010_tile_6 = SSH_OBP_monthly_2010.isel(tile=6)
memory_footp += memory_footprint(SSH_OBP_2010_tile_6)
end_time = time.time()
print('Merge first then subset: ' + str(end_time - start_time) + ' sec elapsed')
print('Memory footprint: ' + str(memory_footp/(2**20)) + ' MB')
# delete datasets from first part to start fresh
del SSH_monthly_2010
del OBP_monthly_2010
del SSH_OBP_monthly_2010
del SSH_OBP_2010_tile_6
# Subset before merging
def subset(ds):
dict_subset = {'tile':6}
ds_subset = ds.isel(dict_subset)
return ds_subset
start_time = time.time()
memory_footp = 0
SSH_2010_tile_6 = xr.open_mfdataset(join(ECCO_dir,'*SSH*MONTHLY*/*_2010-??_*.nc'),preprocess=subset)
memory_footp += memory_footprint(SSH_2010_tile_6)
OBP_2010_tile_6 = xr.open_mfdataset(join(ECCO_dir,'*OBP*MONTHLY*/*_2010-??_*.nc'),preprocess=subset)
memory_footp += memory_footprint(OBP_2010_tile_6)
SSH_OBP_2010_tile_6 = xr.merge([SSH_2010_tile_6,OBP_2010_tile_6])
memory_footp += memory_footprint(SSH_OBP_2010_tile_6)
end_time = time.time()
print('Subset first then merge: ' + str(end_time - start_time) + ' sec elapsed')
print('Memory footprint: ' + str(memory_footp/(2**20)) + ' MB')
Merge first then subset: 11.381447553634644 sec elapsed
Memory footprint: 111.52811431884766 MB
Subset first then merge: 8.979552745819092 sec elapsed
Memory footprint: 30.84374237060547 MB
We can see that subsetting individual datasets before concatenating and merging saves only a little bit of time, but a lot in terms of memory footprint. This matters when dealing with much larger datasets.
[15]:
SSH_OBP_2010_tile_6
[15]:
<xarray.Dataset>
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, time: 12, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
tile int32 6
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
... ...
YC (j, i) float32 dask.array<chunksize=(90, 90), meta=np.ndarray>
XG (j_g, i_g) float32 dask.array<chunksize=(90, 90), meta=np.ndarray>
YG (j_g, i_g) float32 dask.array<chunksize=(90, 90), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (j, i, nb) float32 dask.array<chunksize=(90, 90, 4), meta=np.ndarray>
YC_bnds (j, i, nb) float32 dask.array<chunksize=(90, 90, 4), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
SSHIBC (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
SSHNOIBC (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
ETAN (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
OBP (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
OBPGMAP (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
PHIBOT (time, j, i) float32 dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3- i: 90
- i_g: 90
- j: 90
- j_g: 90
- time: 12
- nv: 2
- nb: 4
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile()int326
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array(6)
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2010-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - XC(j, i)float32dask.array<chunksize=(90, 90), meta=np.ndarray>
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 31.64 kiB 31.64 kiB Shape (90, 90) (90, 90) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray - YC(j, i)float32dask.array<chunksize=(90, 90), meta=np.ndarray>
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 31.64 kiB 31.64 kiB Shape (90, 90) (90, 90) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray - XG(j_g, i_g)float32dask.array<chunksize=(90, 90), meta=np.ndarray>
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
Array Chunk Bytes 31.64 kiB 31.64 kiB Shape (90, 90) (90, 90) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray - YG(j_g, i_g)float32dask.array<chunksize=(90, 90), meta=np.ndarray>
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
Array Chunk Bytes 31.64 kiB 31.64 kiB Shape (90, 90) (90, 90) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray - time_bnds(time, nv)datetime64[ns]dask.array<chunksize=(1, 2), meta=np.ndarray>
- comment :
- Start and end times of averaging period.
- coverage_content_type :
- coordinate
- long_name :
- time bounds of averaging period
Array Chunk Bytes 192 B 16 B Shape (12, 2) (1, 2) Count 98 Tasks 12 Chunks Type datetime64[ns] numpy.ndarray - XC_bnds(j, i, nb)float32dask.array<chunksize=(90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
Array Chunk Bytes 126.56 kiB 126.56 kiB Shape (90, 90, 4) (90, 90, 4) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray - YC_bnds(j, i, nb)float32dask.array<chunksize=(90, 90, 4), meta=np.ndarray>
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
Array Chunk Bytes 126.56 kiB 126.56 kiB Shape (90, 90, 4) (90, 90, 4) Count 67 Tasks 1 Chunks Type float32 numpy.ndarray
- SSH(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - SSHIBC(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- The inverted barometer (IB) correction to sea surface height due to atmospheric pressure loading
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Not an SSH itself, but a correction to model sea level anomaly (ETAN) required to account for the static part of sea surface displacement by atmosphere pressure loading: SSH = SSHNOIBC - SSHIBC. Note: Use SSH for model-data comparisons with altimetry data products that DO apply the IB correction and SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -0.30144819617271423
- valid_max :
- 0.5248842239379883
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - SSHNOIBC(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Sea surface height anomaly without the inverted barometer (IB) correction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Sea surface height anomaly above the geoid without the inverse barometer (IB) correction, suitable for comparisons with altimetry sea surface height data products that do NOT apply the inverse barometer (IB) correction. Note: SSHNOIBC is calculated by correcting model sea level anomaly ETAN for two effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). In ECCO Version 4 Release 4 the model is forced with atmospheric pressure loading. SSHNOIBC does not correct for the static part of the effect of atmosphere pressure loading on sea surface height (the so-called inverse barometer (IB) correction). Use SSH for comparisons with altimetry data products that DO apply the IB correction.
- valid_min :
- -1.6654272079467773
- valid_max :
- 1.4550364017486572
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - ETAN(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Model sea level anomaly
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- Model sea level anomaly WITHOUT corrections for global mean density (steric) changes, inverted barometer effect, or volume displacement due to submerged sea-ice and snow . Note: ETAN should NOT be used for comparisons with altimetry data products because ETAN is NOT corrected for (a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) nor (b) sea level displacement due to submerged sea-ice and snow (see sIceLoad). These corrections ARE made for the variables SSH and SSHNOIBC.
- valid_min :
- -8.304216384887695
- valid_max :
- 1.460192084312439
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - OBP(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Ocean bottom pressure given as equivalent water thickness
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- OBP excludes the contribution from global mean atmospheric pressure and is therefore suitable for comparisons with GRACE data products. OBP is calculated as follows. First, we calculate ocean hydrostatic bottom pressure anomaly, PHIBOT, with PHIBOT = p_b/rhoConst - gH(t), where p_b = model ocean hydrostatic bottom pressure, rhoConst = reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and H(t) is model depth at time t. Then, OBP = PHIBOT/g + corrections for i) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH) and ii) global mean atmospheric pressure variations. Use OBP for comparisons with ocean bottom pressure data products that have been corrected for global mean atmospheric pressure variations. GRACE data typically ARE corrected for global mean atmospheric pressure variations. In contrast, ocean bottom pressure gauge data typically ARE NOT corrected for global mean atmospheric pressure variations.
- valid_min :
- -1.7899188995361328
- valid_max :
- 72.07011413574219
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - OBPGMAP(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Ocean bottom pressure given as equivalent water thickness, includes global mean atmospheric pressure
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- OBPGMAP includes the contribution from global mean atmospheric pressure and is therefore suitable for comparisons with ocean bottom pressure gauge data products. OBPGMAP is calculated as follows. First, we calculate ocean hydrostatic bottom pressure anomaly, PHIBOT, with PHIBOT = p_b/rhoConst - gH(t), where p_b = model ocean hydrostatic bottom pressure, rhoConst = reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and H(t) is model depth at time t. Then, OBPGMAP= PHIBOT/g + corrections for global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH). Use OBPGMAP for comparisons with ocean bottom pressure data products that have NOT been corrected for global mean atmospheric pressure variations. GRACE data typically ARE corrected for global mean atmospheric pressure variations. In contrast, ocean bottom pressure gauge data typically ARE NOT corrected for global mean atmospheric pressure variations.
- valid_min :
- 8.223706245422363
- valid_max :
- 82.08540344238281
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray - PHIBOT(time, j, i)float32dask.array<chunksize=(1, 90, 90), meta=np.ndarray>
- long_name :
- Ocean hydrostatic bottom pressure anomaly
- units :
- m2 s-2
- coverage_content_type :
- modelResult
- comment :
- PHIBOT = p_b / rhoConst - g H(t), where p_b = hydrostatic ocean bottom pressure, rhoConst = reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and H(t) is model depth at time t. Units: p:[kg m-1 s-2], rhoConst:[kg m-3], g:[m s-2], H(t):[m]. Note: includes atmospheric pressure loading. PHIBOT accounts for the model's time-varying grid cell thickness (z* coordinate system). PHIBOT is NOT corrected for global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), and therefore should NOT be used for comparisons with ocean bottom pressure data. Instead, see OBPGMAP and OBP.
- valid_min :
- 80.7757568359375
- valid_max :
- 805.1843872070312
Array Chunk Bytes 379.69 kiB 31.64 kiB Shape (12, 90, 90) (1, 90, 90) Count 48 Tasks 12 Chunks Type float32 numpy.ndarray
- acknowledgement :
- This research was carried out by the Jet Propulsion Laboratory, managed by the California Institute of Technology under a contract with the National Aeronautics and Space Administration.
- author :
- Ian Fenty and Ou Wang
- cdm_data_type :
- Grid
- comment :
- Fields provided on the curvilinear lat-lon-cap 90 (llc90) native grid used in the ECCO model. SSH (dynamic sea surface height) = SSHNOIBC (dynamic sea surface without the inverse barometer correction) - SSHIBC (inverse barometer correction). The inverted barometer correction accounts for variations in sea surface height due to atmospheric pressure variations. Note: ETAN is model sea level anomaly and should not be compared with satellite altimetery products, see SSH and ETAN for more details.
- Conventions :
- CF-1.8, ACDD-1.3
- coordinates_comment :
- Note: the global 'coordinates' attribute describes auxillary coordinates.
- creator_email :
- ecco-group@mit.edu
- creator_institution :
- NASA Jet Propulsion Laboratory (JPL)
- creator_name :
- ECCO Consortium
- creator_type :
- group
- creator_url :
- https://ecco-group.org
- date_created :
- 2020-12-16T18:07:27
- date_issued :
- 2020-12-16T18:07:27
- date_metadata_modified :
- 2021-03-15T21:56:12
- date_modified :
- 2021-03-15T21:56:12
- geospatial_bounds_crs :
- EPSG:4326
- geospatial_lat_max :
- 90.0
- geospatial_lat_min :
- -90.0
- geospatial_lat_resolution :
- variable
- geospatial_lat_units :
- degrees_north
- geospatial_lon_max :
- 180.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_resolution :
- variable
- geospatial_lon_units :
- degrees_east
- history :
- Inaugural release of an ECCO Central Estimate solution to PO.DAAC
- id :
- 10.5067/ECL5M-SSH44
- institution :
- NASA Jet Propulsion Laboratory (JPL)
- instrument_vocabulary :
- GCMD instrument keywords
- keywords :
- EARTH SCIENCE > OCEANS > SEA SURFACE TOPOGRAPHY > SEA SURFACE HEIGHT, EARTH SCIENCE SERVICES > MODELS > EARTH SCIENCE REANALYSES/ASSIMILATION MODELS
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- license :
- Public Domain
- metadata_link :
- https://cmr.earthdata.nasa.gov/search/collections.umm_json?ShortName=ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
- naming_authority :
- gov.nasa.jpl
- platform :
- ERS-1/2, TOPEX/Poseidon, Geosat Follow-On (GFO), ENVISAT, Jason-1, Jason-2, CryoSat-2, SARAL/AltiKa, Jason-3, AVHRR, Aquarius, SSM/I, SSMIS, GRACE, DTU17MDT, Argo, WOCE, GO-SHIP, MEOP, Ice Tethered Profilers (ITP)
- platform_vocabulary :
- GCMD platform keywords
- processing_level :
- L4
- product_name :
- SEA_SURFACE_HEIGHT_mon_mean_2010-01_ECCO_V4r4_native_llc0090.nc
- product_time_coverage_end :
- 2018-01-01T00:00:00
- product_time_coverage_start :
- 1992-01-01T12:00:00
- product_version :
- Version 4, Release 4
- program :
- NASA Physical Oceanography, Cryosphere, Modeling, Analysis, and Prediction (MAP)
- project :
- Estimating the Circulation and Climate of the Ocean (ECCO)
- publisher_email :
- podaac@podaac.jpl.nasa.gov
- publisher_institution :
- PO.DAAC
- publisher_name :
- Physical Oceanography Distributed Active Archive Center (PO.DAAC)
- publisher_type :
- institution
- publisher_url :
- https://podaac.jpl.nasa.gov
- references :
- ECCO Consortium, Fukumori, I., Wang, O., Fenty, I., Forget, G., Heimbach, P., & Ponte, R. M. 2020. Synopsis of the ECCO Central Production Global Ocean and Sea-Ice State Estimate (Version 4 Release 4). doi:10.5281/zenodo.3765928
- source :
- The ECCO V4r4 state estimate was produced by fitting a free-running solution of the MITgcm (checkpoint 66g) to satellite and in situ observational data in a least squares sense using the adjoint method
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- summary :
- This dataset provides monthly-averaged dynamic sea surface height and model sea level anomaly on the lat-lon-cap 90 (llc90) native model grid from the ECCO Version 4 Release 4 (V4r4) ocean and sea-ice state estimate. Estimating the Circulation and Climate of the Ocean (ECCO) state estimates are dynamically and kinematically-consistent reconstructions of the three-dimensional, time-evolving ocean, sea-ice, and surface atmospheric states. ECCO V4r4 is a free-running solution of a global, nominally 1-degree configuration of the MIT general circulation model (MITgcm) that has been fit to observations in a least-squares sense. Observational data constraints used in V4r4 include sea surface height (SSH) from satellite altimeters [ERS-1/2, TOPEX/Poseidon, GFO, ENVISAT, Jason-1,2,3, CryoSat-2, and SARAL/AltiKa]; sea surface temperature (SST) from satellite radiometers [AVHRR], sea surface salinity (SSS) from the Aquarius satellite radiometer/scatterometer, ocean bottom pressure (OBP) from the GRACE satellite gravimeter; sea-ice concentration from satellite radiometers [SSM/I and SSMIS], and in-situ ocean temperature and salinity measured with conductivity-temperature-depth (CTD) sensors and expendable bathythermographs (XBTs) from several programs [e.g., WOCE, GO-SHIP, Argo, and others] and platforms [e.g., research vessels, gliders, moorings, ice-tethered profilers, and instrumented pinnipeds]. V4r4 covers the period 1992-01-01T12:00:00 to 2018-01-01T00:00:00.
- time_coverage_duration :
- P1M
- time_coverage_end :
- 2010-02-01T00:00:00
- time_coverage_resolution :
- P1M
- time_coverage_start :
- 2010-01-01T00:00:00
- title :
- ECCO Sea Surface Height - Monthly Mean llc90 Grid (Version 4 Release 4)
- uuid :
- 9ce7afa6-400c-11eb-ab45-0cc47a3f49c3
Opening and handling very large ECCOv4 datasets using Dask¶
We can load the entire ECCOv4 solution into our workspace and perform calcuations with the entire solution thanks to the amazing Dask library implemented in the xarray package. Why is this useful? Because it is unlikely that the machine you are working on has enough RAM to load the entire ECCOv4 solution at one time. By using Dask we can virtually load all of the ECCO fields into memory. Dask even enables us to do calculations with these fields even though the entire data is
never stored in memory.
Here is some more information about these features from the Dask website, https://docs.dask.org/
Larger-than-memory: Lets you work on datasets that are larger than your available memory by breaking up your array into many small pieces, operating on those pieces in an order that minimizes the memory footprint of your computation, and effectively streaming data from disk.*
Blocked Algorithms: Perform large computations by performing many smaller computations
Finally, in cluster environments Dask can distribute computations across many cores to speed up large redundant calculation.
An in-depth description of Dask is outside the scope of this tutorial. For the moment, let’s load several monthly-mean variables for two years with and without Dask. Then we’ll load the monthly-mean fields for three years using Dask. Without Dask these fields will be loaded into memory. With Dask we will only load a minimum of the Datasets, the Dimensions and Coordinates. At the end we’ll compare their times to load and memory footprints.
Example 1: Load two years monthly-mean ECCO fields into memory without Dask¶
The .load() suffix appended to the end of these load commands is a command to fully load fields into memory. Without it, Dask only virtually loads the fields.
[16]:
# list of shortnames to load datasets of
shortnames_list = ['ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4','ECCO_L4_OBP_LLC0090GRID_MONTHLY_V4R4',\
'ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4']
years_to_load = [2010,2011,2012]
# define function to open multiple datasets and years
def open_multi_datasets_years(shortnames_list,years_to_load):
for count_short,shortname in enumerate(shortnames_list):
for count_year,year in enumerate(years_to_load):
if count_year == 0:
ds_curr = xr.open_mfdataset(join(ECCO_dir,shortname,'*_'+str(year)+'-??_*.nc'))
else:
ds_new = xr.open_mfdataset(join(ECCO_dir,shortname,'*_'+str(year)+'-??_*.nc'))
ds_curr = xr.concat([ds_curr,ds_new],dim='time')
if count_short == 0:
ds = ds_curr
if count_short > 0:
ds = xr.merge([ds,ds_curr])
return ds
# load fields into memory without Dask
t_0 = time.time()
ds = open_multi_datasets_years(shortnames_list,years_to_load)
ds_no_dask = ds.load()
delta_t_3_yrs_no_dask = time.time() - t_0
print(str(delta_t_3_yrs_no_dask) + ' sec elapsed')
25.378690481185913 sec elapsed
Example 2: Open three years of monthly-mean ECCO fields using Dask¶
This time we will omit the .load() suffix and use Dask.
[17]:
t_0 = time.time()
ds = open_multi_datasets_years(shortnames_list,years_to_load)
ds_with_dask = ds
delta_t_3_yrs_with_dask = time.time() - t_0
print(str(delta_t_3_yrs_with_dask) + ' sec elapsed')
11.893571615219116 sec elapsed
Results¶
Now we examine the time it took to load these fields and the comparative memory footprints
[18]:
print ('loaded 3 years without dask in ', delta_t_3_yrs_no_dask, ' sec')
print ('loaded 3 years with_dask in ', delta_t_3_yrs_with_dask, ' sec')
loaded 3 years without dask in 25.378690481185913 sec
loaded 3 years with_dask in 11.893571615219116 sec
The real advantage of using Dask comes when examining the size of these objects in memory.
[19]:
# Estimate the memory footprint in MB
s = memory_footprint(ds_no_dask)
print('ds_no_dask : ', np.round(s/2**20), 'mb')
s = memory_footprint(ds_with_dask)
print('ds_with_dask : ', np.round(s/2**20), 'mb')
ds_no_dask : 1566.0 mb
ds_with_dask : 25.0 mb
Opening/viewing datasets using Dask uses much less memory than loading the full dataset with .load(). Dask uses this “lazy” approach to handling datasets, deferring loading fields into memory and performing calculations until the last possible moment. Combined with Dask’s chunking of datasets, this means that computations can be done even on data spanning much larger sizes than your machine’s RAM.
Summary¶
Now you know efficient ways to open and view the contents of ECCOv4 NetCDF files, including how to concatenate, merge, and subset across multiple times and datasets.
Using Dask we showed that one can prepare a work environment where ECCO model variables are accessible for calculations even without fully loading the fields into memory.