Coordinates and Dimensions of ECCOv4 NetCDF files¶
Objectives¶
Introduce the student to the idea that the ECCO v4 NetCDF fields have coordinate labels which shows where these fields are on the Arakawa-C grid.
Introduction¶
The ECCOv4 files are provided as NetCDF files. The file you have may look a little different than the ones shown here because we have been working hard at improving how what exactly goes into our NetCDF files.
Before you begin, make sure you have the monthly mean temperature/salinity files for 2010 downloaded. If you have done the previous tutorial about Dataset and DataArray objects, you already have these; if not, you should run at least the first code cell of that tutorial before this one.
As we showed in the previous tutorial, we can use the open_mfdataset method from xarray to load multiple NetCDF files into Python as a Dataset object. open_mfdataset is very convenient because it automatically parses and concatenates NetCDF files, constructing a Dataset object using all of the dimensions, coordinates, variables, and metadata information.
In the last tutorial we analyzed the contents of the ECCOv4 monthly mean potential temperature and salinity files for the year 2010. Let’s load these files up again as the Dataset object theta_dataset.
[1]:
import numpy as np
import xarray as xr
import sys
import matplotlib.pyplot as plt
%matplotlib inline
import json
[2]:
## Import the ecco_v4_py library into Python
## =========================================
## -- If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. The code below adds the ECCOv4-py
## folder under the user's home directory to the path.
from os.path import expanduser,join,isdir
import sys
user_home_dir = expanduser('~')
ecco_v4_py_dir = join(user_home_dir,'ECCOv4-py')
if isdir(ecco_v4_py_dir):
sys.path.append(ecco_v4_py_dir)
import ecco_v4_py as ecco
[3]:
## Load temp/sal monthly mean files for 2010
## ================
# indicate whether you are working in a cloud instance (True if yes, False otherwise)
incloud_access = False
## change ECCO_dir as needed
ECCO_dir = join(user_home_dir,'Downloads','ECCO_V4r4_PODAAC')
ShortNames_list = ["ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
if incloud_access == True:
from ecco_s3_retrieve import ecco_podaac_s3_get_diskaware
# retrieve files (download to instance if there is sufficient storage)
files_nested_list = ecco_podaac_s3_get_diskaware(ShortNames=ShortNames_list,\
StartDate='2010-01',EndDate='2010-12',\
max_avail_frac=0.5,\
download_root_dir=ECCO_dir)
# load file into workspace
theta_dataset = xr.open_mfdataset(files_nested_list[ShortNames_list[0]],parallel=True,\
data_vars='minimal',coords='minimal',compat='override')
else:
curr_dir = join(ECCO_dir,ShortNames_list[0])
import glob
# find files on disk (assumes that they were downloaded in the last tutorial)
files_to_load = list(glob.glob(join(curr_dir,'*2010*nc')))
# load file into workspace
theta_dataset = xr.open_mfdataset(files_to_load, parallel=True,\
data_vars='minimal',coords='minimal',compat='override')
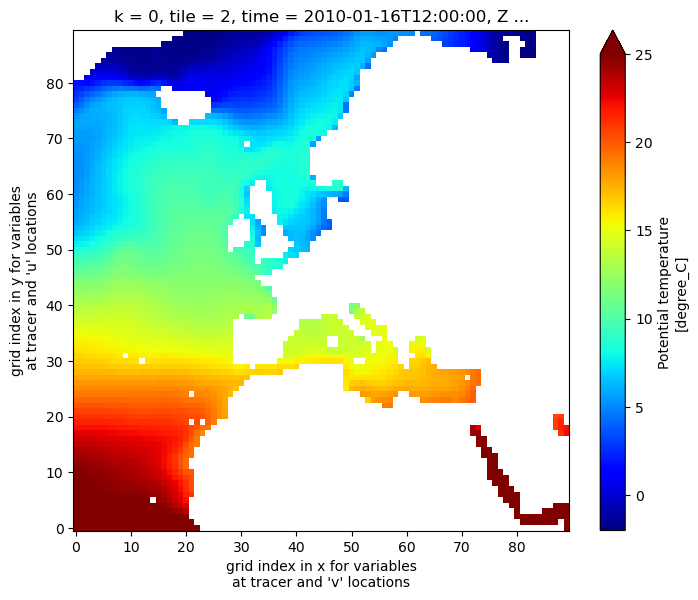
Before we get started, plot the temperature field at the surface layer (k=0) for tile 2, NE Atlantic..
Note :: Don’t worry about the complicated looking code below, w’ll cover plotting later
[4]:
fig=plt.figure(figsize=(8, 6.5))
theta_dataset.THETA.isel(k=0,tile=2,time=0).plot(vmin=-2, vmax=25, cmap='jet')
[4]:
<matplotlib.collections.QuadMesh at 0x7f721d316690>
The Dimensions and Coordinates of THETA¶
Let’s take a closer look at what is inside this dataset. We suppress the metadata (attrs) just to reduce how much is printed to the screen.
[5]:
theta_dataset.attrs = []
theta_dataset
[5]:
<xarray.Dataset> Size: 510MB
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50,
k_p1: 51, tile: 13, time: 12, nv: 2, nb: 4)
Coordinates: (12/22)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 200B 0 1 2 3 4 5 6 7 8 ... 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
Zl (k_l) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] 192B dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Z_bnds (k, nv) float32 400B dask.array<chunksize=(50, 2), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
THETA (time, k, tile, j, i) float32 253MB dask.array<chunksize=(1, 25, 7, 45, 45), meta=np.ndarray>
SALT (time, k, tile, j, i) float32 253MB dask.array<chunksize=(1, 25, 7, 45, 45), meta=np.ndarray>Dimensions¶
theta_dataset shows 12 dimensions; recall that Dataset objects are containers and so it lists all of the unique dimensions of the variables it is storing. theta_dataset is storing two Data variables, THETA and SALT. We see that these data variables are five dimensional from the dimension names within the parentheses, for example: ~~~ THETA (time, tile, k, j, i) float32 0.0 0.0 0.0 0.0 … 0.0 0.0 0.0 0.0 ~~~ Examining the coordinates of the Dataset, we find that all of them
have some combination of the 12 dimensions. Note nv is not a spatial or temporal dimension per se. It is a kind of dummy dimension of length 2 for the coordiante time_bnds which has both a starting and ending time for each one averaging period. The same is true of nb which has length 4 and is used by XC_bnds and YC_bnds to store coordinates for the 4 corners of each tracer grid cell.
Coordinates¶
Dimension Coordinates¶
Beyond having three spatial dimensions theta_dataset also has coordinates in the i, j, and k directions. The most basic coordinates are 1D vectors, one for each dimension, which contain indices for the array. Let us call these basic coordinates, dimension coordinates, whose names are indicated either by an asterisk or boldface in the dataset summary above. Following Python convention, we use 0 as the first index of dimension coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* k_l (k_l) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* k_p1 (k_p1) int32 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49 50
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
Let’s examine the centered dimension coordinates more closely, i.e., (i, j, k, tile, time). The latter part of this tutorial will discuss the spatial structure of dimension coordinates that are not centered on a tracer grid cell, e.g., i_g, k_u.
Dimension Coordinate i¶
[6]:
theta_dataset.i
[6]:
<xarray.DataArray 'i' (i: 90)> Size: 360B
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89],
dtype=int32)
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
Attributes:
axis: X
long_name: grid index in x for variables at tracer and 'v' l...
swap_dim: XC
comment: In the Arakawa C-grid system, tracer (e.g., THETA...
coverage_content_type: coordinatei is an array of integers from 0 to 89 indicating the x_grid_index along this tile’s X axis.
Dimension Coordinate j¶
[7]:
theta_dataset.j
[7]:
<xarray.DataArray 'j' (j: 90)> Size: 360B
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89],
dtype=int32)
Coordinates:
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
Attributes:
axis: Y
long_name: grid index in y for variables at tracer and 'u' l...
swap_dim: YC
comment: In the Arakawa C-grid system, tracer (e.g., THETA...
coverage_content_type: coordinatej is an array of integers from 0 to 89 indicating the y_grid_index along this tile’s Y axis.
Dimension Coordinate k¶
[8]:
theta_dataset.k
[8]:
<xarray.DataArray 'k' (k: 50)> Size: 200B
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49], dtype=int32)
Coordinates:
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
Z (k) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
Attributes:
axis: Z
long_name: grid index in z for tracer variables
swap_dim: Z
coverage_content_type: coordinatek is an array of integers from 0 to 49 indicating the z_grid_index along this tile’s Z axis.
Dimension Coordinate tile¶
[9]:
theta_dataset.tile
[9]:
<xarray.DataArray 'tile' (tile: 13)> Size: 52B
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12], dtype=int32)
Coordinates:
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
Attributes:
long_name: lat-lon-cap tile index
comment: The ECCO V4 horizontal model grid is divided into...
coverage_content_type: coordinatetile is an array of integers from 0 to 12, one for each tile of the lat-lon-cap grid.
Dimension Coordinate time¶
[10]:
theta_dataset.time
[10]:
<xarray.DataArray 'time' (time: 12)> Size: 96B
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000',
'2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000',
'2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000',
'2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000',
'2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000',
'2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 96B 2010-01-16T12:00:00 ... 2010-12-16T12:...
Attributes:
long_name: center time of averaging period
axis: T
bounds: time_bnds
coverage_content_type: coordinate
standard_name: timeIn this file the time coordinate indicates the center time of the averaging period. Recall that we loaded the monthly-mean THETA fields for 2010, so the center time of the averaging periods are the middle of each month in 2010.
Other Coordinates¶
Notice some coordinates do not have an “*” in front of their names:
Coordinates:
XC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=n...
YC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=n...
XG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=n...
YG (tile, j_g, i_g) float32 dask.array<chunksize=(13, 90, 90), meta=n...
Z (k) float32 dask.array<chunksize=(50,), meta=np.ndar...
Zp1 (k_p1) float32 dask.array<chunksize=(51,), meta=np.ndar...
Zu (k_u) float32 dask.array<chunksize=(50,), meta=np.ndar...
Zl (k_l) float32 dask.array<chunksize=(50,), meta=np.ndar...
time_bnds (time, nv) datetime64[ns] dask.array<chunksize=(1, 2), meta=np.nda...
XC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta...
YC_bnds (tile, j, i, nb) float32 dask.array<chunksize=(13, 90, 90, 4), meta...
Z_bnds (k, nv) float32 dask.array<chunksize=(50, 2), meta=np.nd...
These are so-called non-dimension coordinates. From the xarray documenation:
1. non-dimension coordinates are variables that [may] contain coordinate data,
but are not a dimension coordinate.
2. They can be multidimensional ... and there is no relationship between
the name of a non-dimension coordinate and the name(s) of its dimension(s).
3. Non-dimension coordinates can be useful for indexing or plotting; ...
Six of these non-dimension coordinates contain horizontal spatial coordinate data:
XC and YC, the longitudes and latitudes of the ‘c’ points (centered on tile, j, and i)
XG and YG, the longitudes and latitudes of the ‘g’ points (the “lower-left” corners of the tracer grid cells, centered on tile, j_g, and i_g)
XC_bnds and YC_bnds, the longitudes and latitudes of all corners of each grid cell (with the extra nb dimension corresponding to the 4 corners of each grid cell)
Five of these non-dimension coordinates contain vertical spatial coordinate data:
Z, the center depth of tracer grid cells (centered on k)
Zp1, the outer depth bounds of the tracer grid cells (centered on k_p1 with length 1 longer than the vertical length of the grid)
Zu, the depth of the bottom of tracer grid cells (centered on k_u). Be careful, you might think Zu and k_u refer to the upper edge of the grid cell, but actually they refer to the upper edge of the k index, i.e., the bottom edge.
Zl, the depth of the top of tracer grid cells (centered on k_l)
Z_bnds, the depth bounds of each tracer grid cell (with the extra nv dimension of length 2 corresponding to 2 vertical bounds)
And one non-dimension coordinate contains time data:
time_bnds, a 1x2 array of calendar dates and times indicating the start and end times of the averaging period of the field
When multiple DataArrays from different times are combined, the dimensions of the merged arrays will expand along the time dimension.
Let’s quickly look at the time_bnds coordinate. The .values attribute provides the content of an xarray variable as a NumPy array, which is useful for examining data values as well as some numerical manipulations not allowed on xarray DataArrays.
[11]:
theta_dataset.time_bnds.values
[11]:
array([['2010-01-01T00:00:00.000000000', '2010-02-01T00:00:00.000000000'],
['2010-02-01T00:00:00.000000000', '2010-03-01T00:00:00.000000000'],
['2010-03-01T00:00:00.000000000', '2010-04-01T00:00:00.000000000'],
['2010-04-01T00:00:00.000000000', '2010-05-01T00:00:00.000000000'],
['2010-05-01T00:00:00.000000000', '2010-06-01T00:00:00.000000000'],
['2010-06-01T00:00:00.000000000', '2010-07-01T00:00:00.000000000'],
['2010-07-01T00:00:00.000000000', '2010-08-01T00:00:00.000000000'],
['2010-08-01T00:00:00.000000000', '2010-09-01T00:00:00.000000000'],
['2010-09-01T00:00:00.000000000', '2010-10-01T00:00:00.000000000'],
['2010-10-01T00:00:00.000000000', '2010-11-01T00:00:00.000000000'],
['2010-11-01T00:00:00.000000000', '2010-12-01T00:00:00.000000000'],
['2010-12-01T00:00:00.000000000', '2011-01-01T00:00:00.000000000']],
dtype='datetime64[ns]')
For time-averaged fields, time_bnds is a 2D array provding the start and end time of each averaging period.
Let’s look at the third record (March, 2010)
[12]:
theta_dataset.time_bnds[2].values
[12]:
array(['2010-03-01T00:00:00.000000000', '2010-04-01T00:00:00.000000000'],
dtype='datetime64[ns]')
We see that the time bounds are 2010-03-01 to 2010-04-01, which make sense.
Having this time-bounds information readily available can be very helpful.
The Dimension Coordinates of the Arakawa C-Grid¶
Dimension coordinates have special meanings. The MITgcm uses the staggered Arakawa-C grid (hereafter c-grid). In c-grid models, variables are staggered in space. Horizontally, variables are associated with three ‘locations’:
tracer cells (e.g. temperature, salinity, density)
the 4 lateral faces of tracer cells (e.g., horizontal velocities and fluxes)
the 4 corners of tracer cells (e.g., vertical component of vorticity field)
Vertically, there are also two ‘locations’:
tracer cells
the 2 top/bottom faces of tracer cells (e.g., vertical velocities and fluxes).
To understand this better, let’s review the geometry of c-grid models.
3D staggering of velocity components.¶
Paraphrasing from https://mitgcm.readthedocs.io/en/latest/algorithm/c-grid.html,
In c-grid models, the components of flow (u,v,w) are staggered in
space such that the zonal component falls on the interface between
tracer cells in the zonal direction. Similarly for the meridional
and vertical directions.
Why the c-grid?¶
The basic algorithm employed for stepping forward the momentum
equations is based on retaining non-divergence of the flow at
all times. This is most naturally done if the components of flow
are staggered in space in the form of an Arakawa C grid...
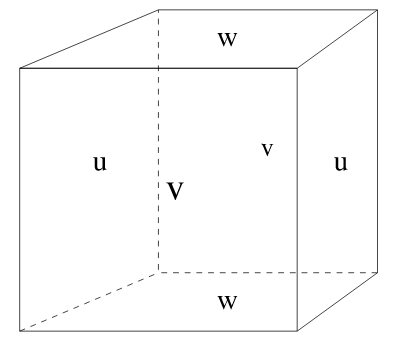
Defining the c-grid coordinate system¶
As shown, variables on Arakawa-C grids (c-grid) are staggered in space. For convenience we define a coordinate system that distinguishes between these different locations.
The c-grid horizontal coordinates¶
In the horizontal, variables can take one of four locations: c, u, v, or g.
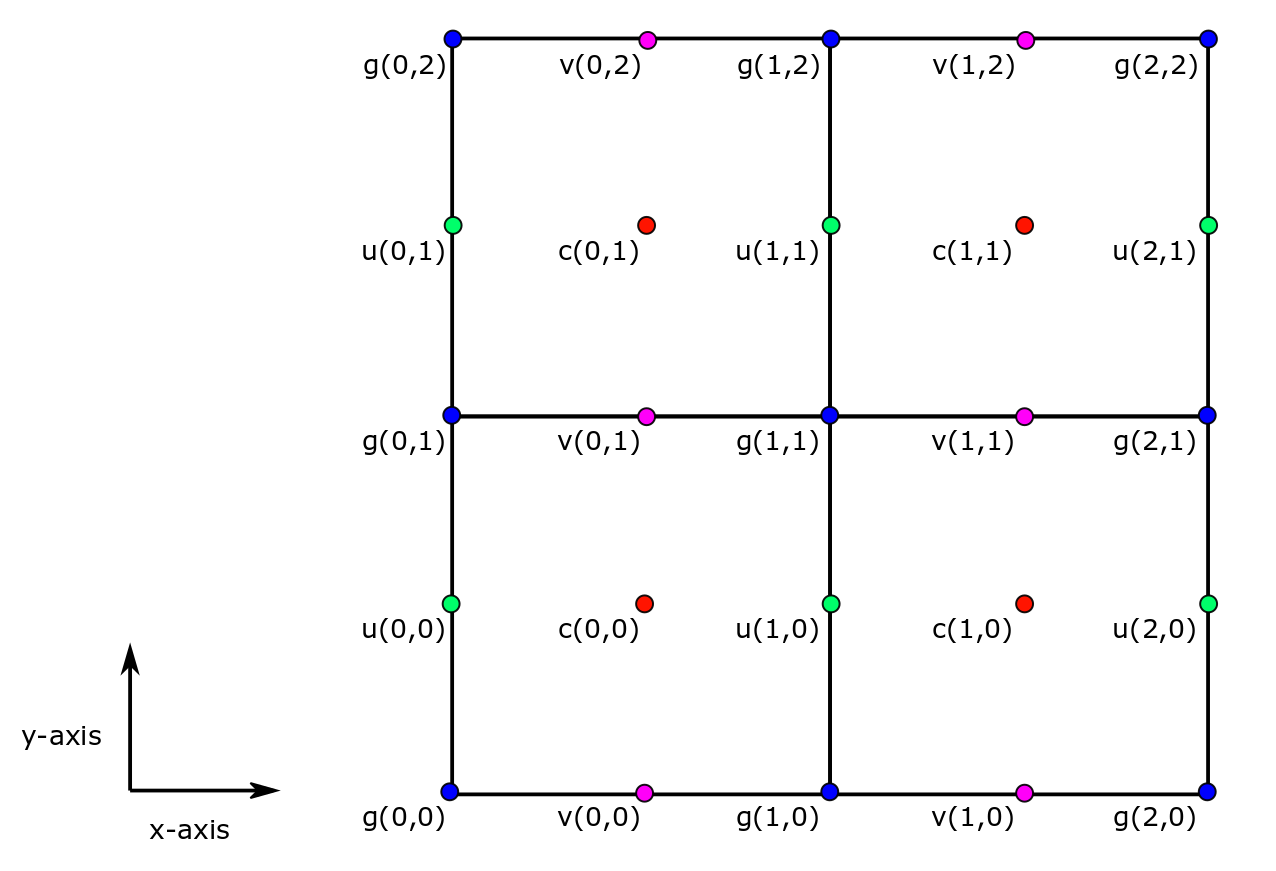
horizontal “c” location¶
Variables associated with tracer cell area or volumetric averages (e.g., temperature, salinity, sea ice thickness, air-sea net heat flux) and variables associated with vertical velocities (e.g., 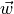 ) are identified with c locations.
) are identified with c locations.
For these variables we define the horizontal dimensions of i and j, corresponding with the model grid 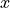 and
and 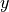 directions, respectively.
directions, respectively.
horizontal “u” location¶
Variables associated with the two lateral sides of tracer cells in the 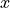 direction are identified with u locations.
direction are identified with u locations.
Define the horizontal dimensions of u variables as i_g and j, corresponding with the model’s 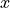 and
and 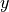 dimensions, respectively.
dimensions, respectively.
Important Note: In the llc90 curvilinear model grid used by ECCOv4, the
direction is NOT same as the geographic zonal (east-west) direction.
horizontal “v” location¶
Variables associated with the two lateral sides of tracer cells in the 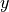 direction are identified with v locations.
direction are identified with v locations.
Define the horizontal dimensions of v variables as i and j_g, corresponding with the model’s 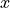 and
and 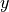 dimensions, respectively.
dimensions, respectively.
Important Note: In the llc90 curvilinear model grid used by ECCOv4, the
direction is NOT same as the geographic meridional (north-south) direction.
horizontal “g” location¶
Variables associated with the horizontal corners of tracer grid cells are identified with g locations.
Define the horizontal dimensions of g variables as i_g and j_g, corresponding with the model’s 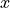 and
and 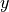 dimensions, respectively.
dimensions, respectively.
The c-grid vertical coordinates¶
In the horizontal, variables can take one of two locations: c and w
vertical “c” location¶
Variables associated with tracer cell volumetric quantities (e.g., temperature, salinity) are identified with c locations.
For these variables we define the vertical dimensions of k which corresponds with the model grid’  direction.
direction.
vertical “w” location¶
Variables associated with the two top/bottom sides of tracer cells in the  direction are identified with w locations.
direction are identified with w locations.
For these variables we define the vertical dimension of :math:`k_u` to indicate the model tracer cell’s upper faces in the 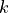 direction, i.e., the bottom faces of the cell.
direction, i.e., the bottom faces of the cell.
Two other vertical dimensions are also used. :math:`k_l` indicates the model tracer cell’s lower faces in the 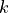 direction (i.e., the top faces of the cell), and
direction (i.e., the top faces of the cell), and 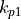 which index all of the upper and lower faces.
which index all of the upper and lower faces.
Note: In ECCOv4 NetCDF files both :math:`k_l`(0) and :math:`k_{p1}`(0) correspond to the same top face of the model tracer grid cell.
All ECCOv4 coordinates¶
Now that we have been oriented to the dimensions and coordinates used by ECCOv4, let’s examine a Dataset that uses all of them, an ECCOv4r4 NetCDF grid file. The grid file for the native LLC90 grid has ShortName ECCO_L4_GEOMETRY_LLC0090GRID_V4R4. It does not have time dimensions, but we can put any StartDate and EndDate between 1992-01-01 and 2018-01-01 into the ecco_podaac_download function (or ecco_podaac_s3_get_diskaware function in the cloud) and it should retrieve the file.
Then the file can be opened; in this case we’ll use open_dataset that loads a single file into memory in our workspace.
[13]:
## download file containing grid parameters and load into workspace
ShortNames_list = ["ECCO_L4_GEOMETRY_LLC0090GRID_V4R4"]
if incloud_access == True:
from ecco_s3_retrieve import ecco_podaac_s3_get_diskaware
# retrieve file (download to instance if there is sufficient storage)
files_nested_list = ecco_podaac_s3_get_diskaware(ShortNames=ShortNames_list,\
StartDate='2010-01',EndDate='2010-12',\
max_avail_frac=0.5,\
download_root_dir=ECCO_dir)
# load file into workspace
grid_dataset = xr.open_dataset(files_nested_list[ShortNames_list[0]])
else:
from ecco_download import ecco_podaac_download
# download grid file
file_to_load = ecco_podaac_download(ShortName=grid_params_shortname,\
StartDate="2010-01-01",EndDate="2010-01-01",\
download_root_dir=ECCO_dir,n_workers=6,force_redownload=False)
# load file into workspace
grid_dataset = xr.open_dataset(files_to_load)
created download directory C:\Users\adelman\Downloads\ECCO_V4r4_PODAAC\ECCO_L4_GEOMETRY_LLC0090GRID_V4R4
{'ShortName': 'ECCO_L4_GEOMETRY_LLC0090GRID_V4R4', 'temporal': '2010-01-01,2010-01-01'}
Total number of matching granules: 1
100%|████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:04<00:00, 4.37s/it]
=====================================
total downloaded: 8.57 Mb
avg download speed: 1.95 Mb/s
Open the ECCOv4r4 grid file:
[14]:
# show contents of grid_dataset
grid_dataset
[14]:
<xarray.Dataset> Size: 89MB
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50,
k_p1: 51, tile: 13, nb: 4, nv: 2)
Coordinates: (12/20)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
... ...
Zp1 (k_p1) float32 204B ...
Zu (k_u) float32 200B ...
Zl (k_l) float32 200B ...
XC_bnds (tile, j, i, nb) float32 2MB ...
YC_bnds (tile, j, i, nb) float32 2MB ...
Z_bnds (k, nv) float32 400B ...
Dimensions without coordinates: nb, nv
Data variables: (12/21)
CS (tile, j, i) float32 421kB ...
SN (tile, j, i) float32 421kB ...
rA (tile, j, i) float32 421kB ...
dxG (tile, j_g, i) float32 421kB ...
dyG (tile, j, i_g) float32 421kB ...
Depth (tile, j, i) float32 421kB ...
... ...
hFacC (k, tile, j, i) float32 21MB ...
hFacW (k, tile, j, i_g) float32 21MB ...
hFacS (k, tile, j_g, i) float32 21MB ...
maskC (k, tile, j, i) bool 5MB ...
maskW (k, tile, j, i_g) bool 5MB ...
maskS (k, tile, j_g, i) bool 5MB ...
Attributes: (12/58)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
references: ECCO Consortium, Fukumori, I., Wang, O.,...
source: The ECCO V4r4 state estimate was produce...
standard_name_vocabulary: NetCDF Climate and Forecast (CF) Metadat...
summary: This dataset provides geometric paramete...
title: ECCO Geometry Parameters for the Lat-Lon...
uuid: 87ff7d24-86e5-11eb-9c5f-f8f21e2ee3e0You’ll notice that the dimensions and coordinates of the grid file are the same as for the potential temperature/salinity files we looked at before, except that there is no time dimension or time-related coordinates. Let’s move on to the Data variables, which in this file are parameters associated with the grid.
Grid Parameters¶
There are 21 data variables in the grid file. These grid parameters describe the model grid geometry, and are essential for quantitative analysis with ECCO output.
axis rotation factors:
CS (tile, j, i) float32 ...
SN (tile, j, i) float32 ...
horizontal distances:
dxG (tile, j_g, i) float32 ...
dyG (tile, j, i_g) float32 ...
dxC (tile, j, i_g) float32 ...
dyC (tile, j_g, i) float32 ...
vertical distances:
drC (k_p1) float32 ...
drF (k) float32 ...
areas:
rA (tile, j, i) float32 ...
rAz (tile, j_g, i_g) float32 ...
rAw (tile, j, i_g) float32 ...
rAs (tile, j_g, i) float32 ...
reference hydrostatic pressure at cell depth:
PHrefC (k) float32 ...
PHrefF (k_p1) float32 ...
partial cell fractions:
hFacC (k, tile, j, i) float32 ...
hFacW (k, tile, j, i_g) float32 ...
hFacS (k, tile, j_g, i) float32 ...
wet/dry boolean masks:
maskC (k, tile, j, i) bool ...
maskW (k, tile, j, i_g) bool ...
maskS (k, tile, j_g, i) bool ...
seafloor depth:
Depth (j, i) float32 ...
Grid parameters are not coordinates in the sense that they help you orient in space or time, but they provide measures of the model grid such as distances and areas. Let’s examine one of these grid geometric variables, dxG:
[15]:
grid_dataset.dxG
[15]:
<xarray.DataArray 'dxG' (tile: 13, j_g: 90, i: 90)> Size: 421kB
[105300 values with dtype=float32]
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
Attributes:
long_name: distance between 'southwest' and 'southeast' corn...
units: m
coordinate: YG XC
coverage_content_type: modelResult
comment: Alternatively, the length of 'south' side of trac...dxG has coordinates tile, j_g and i which means that it is a v location variable. dxG is the horizontal distance between 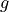 points (tracer cell corners) in the tile’s
points (tracer cell corners) in the tile’s 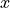 direction.
direction.
For reference, see the chart below from the MITgcm documentation, Figure 2.6
dxG = 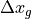 in subfigure (a) below:
in subfigure (a) below:
Figure 2.6 Staggering of horizontal grid descriptors (lengths and areas). The grid lines indicate the tracer cell boundaries and are the reference grid for all panels. a) The area of a tracer cell, 𝐴𝑐, is bordered by the lengths Δ𝑥𝑔 and Δ𝑦𝑔. b) The area of a vorticity cell, 𝐴𝜁, is bordered by the lengths Δ𝑥𝑐 and Δ𝑦𝑐. c) The area of a u cell, 𝐴𝑤, is bordered by the lengths Δ𝑥𝑣 and Δ𝑦𝑓. d) The area of a v cell, 𝐴𝑠, is bordered by the lengths Δ𝑥𝑓 and Δ𝑦𝑢.
Dimensions and Coordinates of Velocities¶
So far we looked at files with THETA and SALT which are  variables (i.e. tracers). Let’s examine UVEL, horizontal velocity in the tile’s
variables (i.e. tracers). Let’s examine UVEL, horizontal velocity in the tile’s 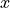 direction. As you’ve probably guessed, UVEL is a
direction. As you’ve probably guessed, UVEL is a 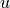 variable:
variable:
Let’s download and open the March 2010 mean horizontal velocity.
[16]:
## download file containing monthly mean ocean velocities for March 2010, and load into workspace
ShortNames_list = ["ECCO_L4_OCEAN_VEL_LLC0090GRID_MONTHLY_V4R4"]
if incloud_access == True:
# retrieve file (download to instance if there is sufficient storage)
files_nested_list = ecco_podaac_s3_get_diskaware(ShortNames=ShortNames_list,\
StartDate='2010-03',EndDate='2010-03',\
max_avail_frac=0.5,\
download_root_dir=ECCO_dir)
# load file into workspace
vel_dataset = xr.open_dataset(files_nested_list[ShortNames_list[0]])
else:
# download velocity file
file_to_load = ecco_podaac_download(ShortName=vel_shortname,\
StartDate='2010-03',EndDate='2010-03',\
download_root_dir=ECCO_dir,n_workers=6,force_redownload=False)
# load file into workspace
vel_dataset = xr.open_dataset(file_to_load)
created download directory C:\Users\adelman\Downloads\ECCO_V4r4_PODAAC\ECCO_L4_OCEAN_VEL_LLC0090GRID_MONTHLY_V4R4
{'ShortName': 'ECCO_L4_OCEAN_VEL_LLC0090GRID_MONTHLY_V4R4', 'temporal': '2010-03-02,2010-03-31'}
Total number of matching granules: 1
100%|████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:04<00:00, 4.90s/it]
=====================================
total downloaded: 30.58 Mb
avg download speed: 6.21 Mb/s
[17]:
# remove attributes from file and look at the file contents
vel_dataset.attrs = []
vel_dataset
[17]:
<xarray.Dataset> Size: 68MB
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50,
k_p1: 51, tile: 13, time: 1, nv: 2, nb: 4)
Coordinates: (12/22)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 200B 0 1 2 3 4 5 6 7 8 ... 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 200B ...
Zl (k_l) float32 200B ...
time_bnds (time, nv) datetime64[ns] 16B ...
XC_bnds (tile, j, i, nb) float32 2MB ...
YC_bnds (tile, j, i, nb) float32 2MB ...
Z_bnds (k, nv) float32 400B ...
Dimensions without coordinates: nv, nb
Data variables:
UVEL (time, k, tile, j, i_g) float32 21MB ...
VVEL (time, k, tile, j_g, i) float32 21MB ...
WVEL (time, k_l, tile, j, i) float32 21MB ...Notice that each of the velocity variables are centered at different locations, and none of them are at the center of the grid cell (k, tile, j, i). UVEL has the i_g dimension, indicating that it is on the edge of each tracer cell along the 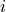 axis.
axis. VVEL has the j_g dimension, indicating that it is on the edge of each tracer cell along the 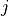 axis.
axis. WVEL has the k_l dimension, indicating that it is on the edge of each tracer cell along the 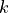 (vertical) axis.
(vertical) axis.
Let’s confirm this by looking at the dimensions and coordinates of each of the velocity files.
UVEL¶
[18]:
vel_dataset.UVEL.dims
[18]:
('time', 'k', 'tile', 'j', 'i_g')
[19]:
vel_dataset.UVEL.coords
[19]:
Coordinates:
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 8B 2010-03-16T12:00:00
Z (k) float32 200B ...
Plot tile 1 time-mean horizontal velocity in the tile’s 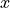 direction, at the top-most model grid cell.
direction, at the top-most model grid cell.
[20]:
fig=plt.figure(figsize=(8, 6.5))
ud_masked = vel_dataset.UVEL.where(grid_dataset.hFacW > 0, np.nan)
ud_masked.isel(k=0,tile=1,time=0).plot(cmap='jet', vmin=-.2,vmax=.2)
[20]:
<matplotlib.collections.QuadMesh at 0x7f71ff018b90>
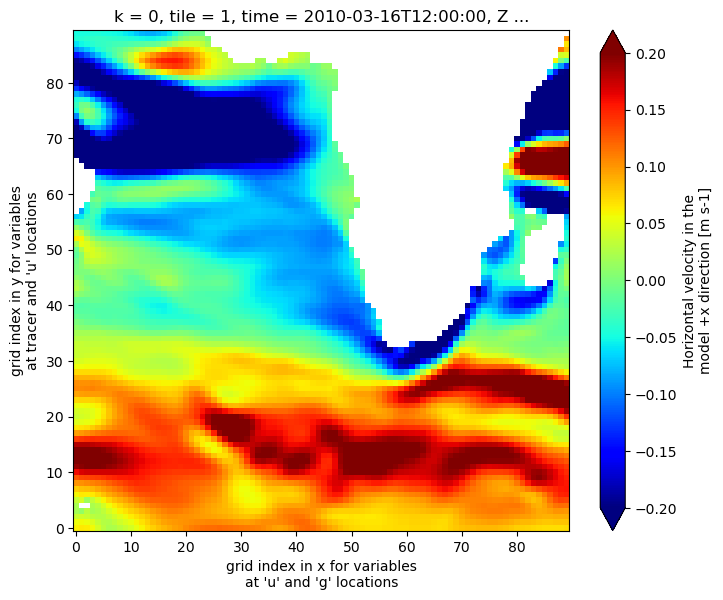
VVEL¶
[21]:
vel_dataset.VVEL.dims
[21]:
('time', 'k', 'tile', 'j_g', 'i')
[22]:
vel_dataset.VVEL.coords
[22]:
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 8B 2010-03-16T12:00:00
Z (k) float32 200B -5.0 -15.0 -25.0 ... -5.461e+03 -5.906e+03
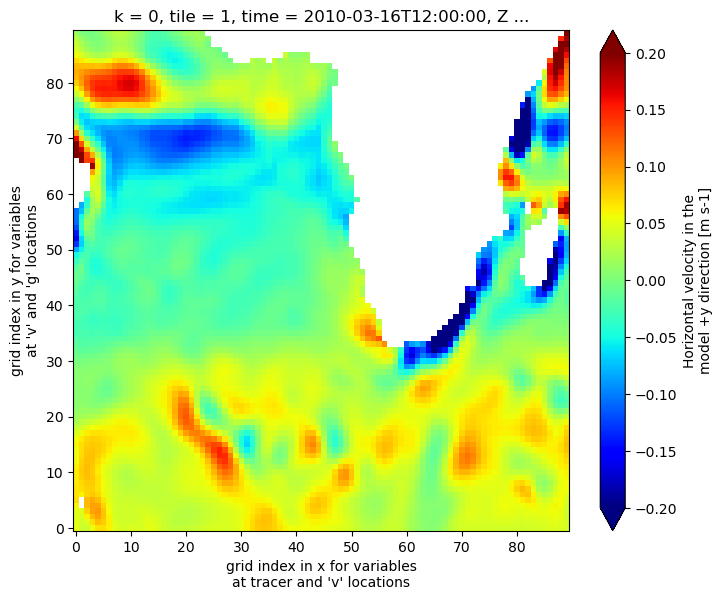
Plot the time-mean horizontal velocity in the tile’s 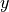 direction, at the top-most model grid cell.
direction, at the top-most model grid cell.
[23]:
fig=plt.figure(figsize=(8, 6.5))
vd_masked = vel_dataset.VVEL.where(grid_dataset.hFacS > 0, np.nan)
vd_masked.isel(k=0,tile=1,time=0).plot(cmap='jet', vmin=-.2,vmax=.2)
[23]:
<matplotlib.collections.QuadMesh at 0x7f71ff0c77d0>
WVEL¶
[24]:
vel_dataset.WVEL.dims
[24]:
('time', 'k_l', 'tile', 'j', 'i')
[25]:
vel_dataset.WVEL.coords
[25]:
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* k_l (k_l) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 8B 2010-03-16T12:00:00
XC (tile, j, i) float32 421kB ...
YC (tile, j, i) float32 421kB ...
Zl (k_l) float32 200B ...
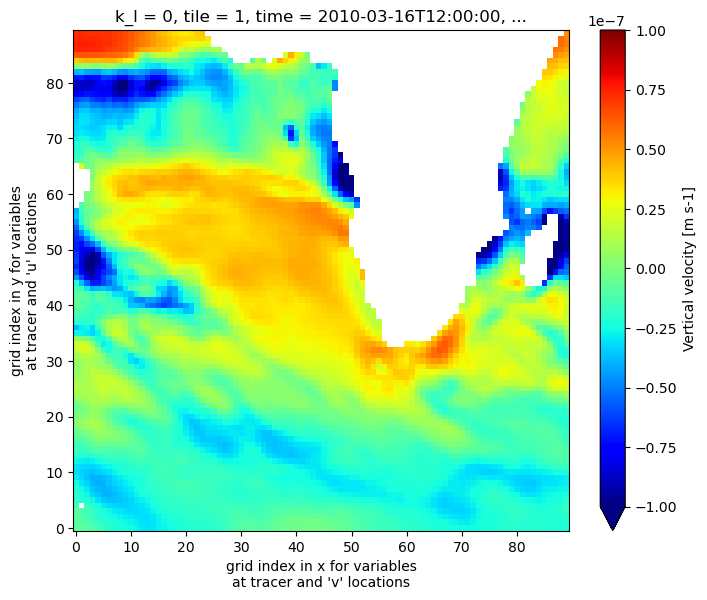
Plot the time-mean vertical velocity on the tile, at the ocean surface. Note that the vertical index we pass to isel is k_l=0 rather than k=0, corresponding to the vertical dimension and coordinate of WVEL.
[26]:
fig=plt.figure(figsize=(8, 6.5))
# use .values attribute below to avoid xarray getting confused about the different vertical dimensions (k_l vs. k)
wd_masked = vel_dataset.WVEL.where(grid_dataset.hFacC.values > 0, np.nan)
wd_masked.isel(k_l=0,tile=1,time=0).plot(cmap='jet', vmin=-1.e-7,vmax=1.e-7)
[26]:
<matplotlib.collections.QuadMesh at 0x7f71fefea0d0>
Summary¶
ECCOv4 variables are on the staggered Arakawa-C grid. Different dimension labels and coordinates are applied to state estimate variables so that one can easily identify where on the c-grid any particular variable is situated.