Combining multiple Datasets¶
Objectives:¶
Show how to combine multiple ECCO v4 state estimate Datasets after loading.
Opening multiple Datasets centered on different coordinates¶
In previous tutorials we’ve loaded single lat-lon-cap NetCDF tile files (granules) for ECCO state estimate variables and model grid parameters. Here we will demonstrate merging Datasets together. Some benefits of merging Datasets include having a tidier workspace and simplifying subsetting operations (e.g., using xarray.isel or xarray.sel as shown in the previous
tutorial).
First, we’ll load three ECCOv4 NetCDF state estimate variables (each centered on different coordinates) as well as the model grid file. For this, you will need to download 2 datasets of monthly mean fields for the year 2010. The ShortNames for the 2 datasets are:
ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
ECCO_L4_OCEAN_3D_TEMPERATURE_FLUX_LLC0090GRID_MONTHLY_V4R4
If you did the previous tutorial you already have the SSH files.
Once you have the required ECCOv4 output downloaded, let’s define our environment.
[1]:
import numpy as np
import xarray as xr
import sys
import matplotlib.pyplot as plt
import json
[2]:
## Import the ecco_v4_py library into Python
## =========================================
#import ecco_v4_py as ecco
## -- If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. For example, if your ecco_v4_py
## files are in /Users/ifenty/ECCOv4-py/ecco_v4_py, then use:
from os.path import join,expanduser
user_home_dir = expanduser('~')
sys.path.append(join(user_home_dir,'ECCOv4-py'))
import ecco_v4_py as ecco
[3]:
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to ~/Downloads/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join(user_home_dir,'Downloads','ECCO_V4r4_PODAAC')
Open c point variable: SSH¶
[4]:
# load dataset containing monthly SSH in 2010
ecco_dataset_A = xr.open_mfdataset(join(ECCO_dir,'*SSH*MONTHLY*','*_2010-??_*.nc'))
to see the data variables in a dataset, use .data_vars:
[5]:
ecco_dataset_A.data_vars
[5]:
Data variables:
SSH (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHNOIBC (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ETAN (time, tile, j, i) float32 dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ecco_dataset_A has four data variables, all having dimensions i, j, tile, and time, which mean that they are centered with respect to the grid cells of the model. The coordinates associated with the SSH variable are:
[6]:
ecco_dataset_A.SSH.coords
[6]:
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
XC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YC (tile, j, i) float32 dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
You can see the coordinates that are also dimensions (dimensional coordinates) have an asterisk. The non-dimensional coordinates XC and YC are not used for indexing, but are very important as they indicate the longitude and latitude respectively associated with the dimensional coordinates.
Open u and v point variables: ADVx_TH and ADVy_TH¶
Now let’s open the ECCOv4 output files containing the horizontal advective fluxes of potential temperature, ADVx_TH and ADVy_TH.
[7]:
# load dataset containing monthly mean 3D temperature fluxes in 2010
ecco_dataset_B = xr.open_mfdataset(join(ECCO_dir,'*3D_TEMPERATURE_FLUX_LLC0090GRID_MONTHLY*','*_2010-??_*.nc'))
ecco_dataset_B.data_vars
[7]:
Data variables:
ADVx_TH (time, k, tile, j, i_g) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
DFxE_TH (time, k, tile, j, i_g) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
ADVy_TH (time, k, tile, j_g, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
DFyE_TH (time, k, tile, j_g, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
ADVr_TH (time, k_l, tile, j, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
DFrE_TH (time, k_l, tile, j, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
DFrI_TH (time, k_l, tile, j, i) float32 dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
ecco_dataset_B has seven data variables! These include three variables (starting with AD) that quantify 3D advection fluxes, and four variables (starting with DF) that quantify 3D diffusive fluxes, including one DFrI_TH that is an implicit flux from the vertical mixing parameterization of the model. In this tutorial we will focus on the two variables that quantify horizontal advective fluxes.
Let’s look at one of these variables, ADVx_TH
[8]:
ecco_dataset_B.ADVx_TH
[8]:
<xarray.DataArray 'ADVx_TH' (time: 12, k: 50, tile: 13, j: 90, i_g: 90)>
dask.array<concatenate, shape=(12, 50, 13, 90, 90), dtype=float32, chunksize=(1, 50, 13, 90, 90), chunktype=numpy.ndarray>
Coordinates:
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
Z (k) float32 dask.array<chunksize=(50,), meta=np.ndarray>
Attributes:
long_name: Lateral advective flux of potential temperature i...
units: degree_C m3 s-1
mate: ADVy_TH
coverage_content_type: modelResult
direction: >0 increases potential temperature (THETA)
comment: Lateral advective flux of potential temperature (...
valid_min: -28231902.0
valid_max: 36523468.0- time: 12
- k: 50
- tile: 13
- j: 90
- i_g: 90
- dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
Array Chunk Bytes 241.01 MiB 20.08 MiB Shape (12, 50, 13, 90, 90) (1, 50, 13, 90, 90) Count 36 Tasks 12 Chunks Type float32 numpy.ndarray - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2010-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - Z(k)float32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
Array Chunk Bytes 200 B 200 B Shape (50,) (50,) Count 55 Tasks 1 Chunks Type float32 numpy.ndarray
- long_name :
- Lateral advective flux of potential temperature in the model +x direction
- units :
- degree_C m3 s-1
- mate :
- ADVy_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +x direction through the 'u' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVx_TH(i_g,j,k) corresponds to +x fluxes through the 'u' face of the tracer cell at (i,j,k). Also, the model +x direction does not necessarily correspond to the geographical east-west direction because the x and y axes of the model's lat-lon-cap (llc) curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -28231902.0
- valid_max :
- 36523468.0
The long_name and the comment tell us that this variable is the advective flux of potential temperature in the model +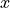 direction. It has dimensional coordinates i_g, j, k, tile, and time. The k dimension indexes depth (which was not a part of the
direction. It has dimensional coordinates i_g, j, k, tile, and time. The k dimension indexes depth (which was not a part of the SSH dataset), and the i_g dimension has replaced the i dimension that SSH had.
Since ADVx_TH has a i_g dimensional coordinate instead of i, we know that the flux is quantified not at the center of each cell, but is offset in the model 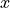 direction. Specifically, the flux is quantified on the left or “west” face of each grid cell. In this context “west” may not actually refer to geographical west, but rather the left side of a grid cell on an axis where i and i_g increase to the right.
direction. Specifically, the flux is quantified on the left or “west” face of each grid cell. In this context “west” may not actually refer to geographical west, but rather the left side of a grid cell on an axis where i and i_g increase to the right.
Now consider ADVy_TH:
[9]:
ecco_dataset_B.ADVy_TH
[9]:
<xarray.DataArray 'ADVy_TH' (time: 12, k: 50, tile: 13, j_g: 90, i: 90)>
dask.array<concatenate, shape=(12, 50, 13, 90, 90), dtype=float32, chunksize=(1, 50, 13, 90, 90), chunktype=numpy.ndarray>
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
Z (k) float32 dask.array<chunksize=(50,), meta=np.ndarray>
Attributes:
long_name: Lateral advective flux of potential temperature i...
units: degree_C m3 s-1
mate: ADVx_TH
coverage_content_type: modelResult
direction: >0 increases potential temperature (THETA)
comment: Lateral advective flux of potential temperature (...
valid_min: -31236064.0
valid_max: 43466144.0- time: 12
- k: 50
- tile: 13
- j_g: 90
- i: 90
- dask.array<chunksize=(1, 50, 13, 90, 90), meta=np.ndarray>
Array Chunk Bytes 241.01 MiB 20.08 MiB Shape (12, 50, 13, 90, 90) (1, 50, 13, 90, 90) Count 36 Tasks 12 Chunks Type float32 numpy.ndarray - i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2010-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - Z(k)float32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
Array Chunk Bytes 200 B 200 B Shape (50,) (50,) Count 55 Tasks 1 Chunks Type float32 numpy.ndarray
- long_name :
- Lateral advective flux of potential temperature in the model +y direction
- units :
- degree_C m3 s-1
- mate :
- ADVx_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +y direction through the 'v' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVy_TH(i,j_g,k) corresponds to +y fluxes through the 'v' face of the tracer cell at (i,j,k). Also, the model +y direction does not necessarily correspond to the geographical north-south direction because the x and y axes of the model's curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -31236064.0
- valid_max :
- 43466144.0
ADVy_TH is the horizontal advective flux of potential temperature in each tile’s 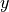 direction. The dimensional coordinates are i, j_g, k, tile, and time. In this case we have the centered
direction. The dimensional coordinates are i, j_g, k, tile, and time. In this case we have the centered 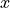 coordinate i but the off-center (shifted)
coordinate i but the off-center (shifted) 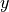 coordinate j_g, which indicates that these fluxes are located on the lower or “south” face of each grid cell—again with the caveat that this does not always correspond to geographical south.
coordinate j_g, which indicates that these fluxes are located on the lower or “south” face of each grid cell—again with the caveat that this does not always correspond to geographical south.
Examining the dimensions and coordinates of these Datasets¶
Each of the three variables we have discussed comprises an xarray DataArray, and each of these DataArray objects has different horizontal dimension labels.
i and j for SSH
i_g and j for ADVx_TH
i and j_g for ADVy_TH
[10]:
# print just the first line of each DataArray's information
print((str(ecco_dataset_A.SSH)).split('\n')[0])
print((str(ecco_dataset_B.ADVx_TH)).split('\n')[0])
print((str(ecco_dataset_B.ADVy_TH)).split('\n')[0])
<xarray.DataArray 'SSH' (time: 12, tile: 13, j: 90, i: 90)>
<xarray.DataArray 'ADVx_TH' (time: 12, k: 50, tile: 13, j: 90, i_g: 90)>
<xarray.DataArray 'ADVy_TH' (time: 12, k: 50, tile: 13, j_g: 90, i: 90)>
Merging multiple Datasets from state estimate variables¶
Using the xarray method merge we can create a single Dataset with multiple DataArrays.
[11]:
# merge together
ecco_dataset_AB = xr.merge([ecco_dataset_A['SSH'], ecco_dataset_B[['ADVx_TH','ADVy_TH']]]).load()
Note: There are multiple syntaxes for selecting individual variables/DataArrays from an
xarraydataset. In the case of SSH above, see that we used bothecco_dataset_A.SSHandecco_dataset_A['SSH'], they indicate the same DataArray. The bracket syntax has the advantage of allowing you to create a subset dataset with multiple variables, by passing a list of the variable names, e.g.,ecco_dataset_B[['ADVx_TH','ADVy_TH']].
Examining the merged Dataset¶
As before, let’s look at the contents of the new merged Dataset
[12]:
ecco_dataset_AB
[12]:
<xarray.Dataset>
Dimensions: (i: 90, j: 90, tile: 13, time: 12, k: 50, i_g: 90, j_g: 90)
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
XC (tile, j, i) float32 -111.6 -111.3 -110.9 ... -99.42 -105.6 -111.9
YC (tile, j, i) float32 -88.24 -88.38 -88.52 ... -88.03 -88.08 -88.1
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
XG (tile, j_g, i_g) float32 -115.0 -115.0 -115.0 ... -102.9 -109.0
YG (tile, j_g, i_g) float32 -88.18 -88.32 -88.46 ... -87.99 -88.02
Z (k) float32 -5.0 -15.0 -25.0 ... -5.039e+03 -5.461e+03 -5.906e+03
Data variables:
SSH (time, tile, j, i) float32 nan nan nan nan nan ... nan nan nan nan
ADVx_TH (time, k, tile, j, i_g) float32 nan nan nan nan ... nan nan nan nan
ADVy_TH (time, k, tile, j_g, i) float32 nan nan nan nan ... nan nan nan nan
Attributes:
long_name: Dynamic sea surface height anomaly
units: m
coverage_content_type: modelResult
standard_name: sea_surface_height_above_geoid
comment: Dynamic sea surface height anomaly above the geoi...
valid_min: -1.8805772066116333
valid_max: 1.4207719564437866- i: 90
- j: 90
- tile: 13
- time: 12
- k: 50
- i_g: 90
- j_g: 90
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2010-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - XC(tile, j, i)float32-111.6 -111.3 ... -105.6 -111.9
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-111.60647 , -111.303 , -110.94285 , ..., 64.791115, 64.80521 , 64.81917 ], [-104.8196 , -103.928444, -102.87706 , ..., 64.36745 , 64.41012 , 64.4524 ], [ -98.198784, -96.788055, -95.14185 , ..., 63.936497, 64.008224, 64.0793 ], ..., [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ]], [[ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], ... [ -40.5 , -40.5 , -40.5 , ..., -40.5 , -40.5 , -40.5 ], [ -39.5 , -39.5 , -39.5 , ..., -39.5 , -39.5 , -39.5 ], [ -38.5 , -38.5 , -38.5 , ..., -38.5 , -38.5 , -38.5 ]], [[-127.5 , -127.5 , -127.5 , ..., -115.850204, -115.50567 , -115.166985], [-126.5 , -126.5 , -126.5 , ..., -115.78025 , -115.464066, -115.153244], [-125.5 , -125.5 , -125.5 , ..., -115.71079 , -115.42275 , -115.139595], ..., [ -40.5 , -40.5 , -40.5 , ..., -101.42989 , -106.83081 , -112.28605 ], [ -39.5 , -39.5 , -39.5 , ..., -100.48844 , -106.24874 , -112.090065], [ -38.5 , -38.5 , -38.5 , ..., -99.42048 , -105.58465 , -111.86579 ]]], dtype=float32) - YC(tile, j, i)float32-88.24 -88.38 ... -88.08 -88.1
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.24259 , -88.382515 , -88.52242 , ..., -80.57383 , -80.50492 , -80.43992 ], [-88.21676 , -88.354485 , -88.49178 , ..., -80.56925 , -80.50038 , -80.43542 ], [-88.164764 , -88.29827 , -88.43065 , ..., -80.55984 , -80.49105 , -80.42617 ], ..., [-58.321964 , -58.321964 , -58.321964 , ..., -58.321964 , -58.321964 , -58.321964 ], [-57.79962 , -57.79962 , -57.79962 , ..., -57.79962 , -57.79962 , -57.79962 ], [-57.271408 , -57.271408 , -57.271408 , ..., -57.271408 , -57.271408 , -57.271408 ]], [[-56.73891 , -56.73891 , -56.73891 , ..., -56.73891 , -56.73891 , -56.73891 ], [-56.2021 , -56.2021 , -56.2021 , ..., -56.2021 , -56.2021 , -56.2021 ], [-55.65936 , -55.65936 , -55.65936 , ..., -55.65936 , -55.65936 , -55.65936 ], ... [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ]], [[-57.271408 , -57.79962 , -58.321964 , ..., -80.36532 , -80.3745 , -80.37896 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.30862 , -80.317726 , -80.32216 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.25618 , -80.26523 , -80.26963 ], ..., [-57.271408 , -57.79962 , -58.321964 , ..., -87.7596 , -87.80198 , -87.822876 ], [-57.271408 , -57.79962 , -58.321964 , ..., -87.89526 , -87.94044 , -87.96276 ], [-57.271408 , -57.79962 , -58.321964 , ..., -88.030365 , -88.07871 , -88.10267 ]]], dtype=float32) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - XG(tile, j_g, i_g)float32-115.0 -115.0 ... -102.9 -109.0
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-115. , -115. , -115. , ..., 65. , 65. , 65. ], [-108.47213 , -107.915245, -107.26045 , ..., 64.56598 , 64.59443 , 64.62261 ], [-101.9954 , -100.91797 , -99.66073 , ..., 64.12406 , 64.18147 , 64.23835 ], ..., [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ]], [[ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], ... [ -41. , -41. , -41. , ..., -41. , -41. , -41. ], [ -40. , -40. , -40. , ..., -40. , -40. , -40. ], [ -39. , -39. , -39. , ..., -39. , -39. , -39. ]], [[-128. , -128. , -128. , ..., -116.06686 , -115.70526 , -115.34945 ], [-127. , -127. , -127. , ..., -115.982254, -115.64933 , -115.32173 ], [-126. , -126. , -126. , ..., -115.89828 , -115.59381 , -115.29422 ], ..., [ -41. , -41. , -41. , ..., -99.28857 , -104.466286, -109.73619 ], [ -40. , -40. , -40. , ..., -98.24634 , -103.74563 , -109.36924 ], [ -39. , -39. , -39. , ..., -97.07265 , -102.928925, -108.95171 ]]], dtype=float32) - YG(tile, j_g, i_g)float32-88.18 -88.32 ... -87.99 -88.02
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.17569 , -88.31587 , -88.45609 , ..., -80.610275, -80.53945 , -80.47247 ], [-88.16335 , -88.30251 , -88.44153 , ..., -80.608 , -80.5372 , -80.470245], [-88.12586 , -88.26202 , -88.397514, ..., -80.601006, -80.530266, -80.46337 ], ..., [-58.58002 , -58.58002 , -58.58002 , ..., -58.58002 , -58.58002 , -58.58002 ], [-58.06183 , -58.06183 , -58.06183 , ..., -58.06183 , -58.06183 , -58.06183 ], [-57.53605 , -57.53605 , -57.53605 , ..., -57.53605 , -57.53605 , -57.53605 ]], [[-57.005695, -57.005695, -57.005695, ..., -57.005695, -57.005695, -57.005695], [-56.471046, -56.471046, -56.471046, ..., -56.471046, -56.471046, -56.471046], [-55.931786, -55.931786, -55.931786, ..., -55.931786, -55.931786, -55.931786], ... [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046]], [[-57.005695, -57.53605 , -58.06183 , ..., -80.38878 , -80.400444, -80.40726 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.33003 , -80.34161 , -80.34837 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.27549 , -80.286995, -80.29371 ], ..., [-57.005695, -57.53605 , -58.06183 , ..., -87.66318 , -87.71477 , -87.7454 ], [-57.005695, -57.53605 , -58.06183 , ..., -87.797325, -87.85214 , -87.884766], [-57.005695, -57.53605 , -58.06183 , ..., -87.93075 , -87.9892 , -88.02409 ]]], dtype=float32) - Z(k)float32-5.0 -15.0 ... -5.906e+03
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([-5.000000e+00, -1.500000e+01, -2.500000e+01, -3.500000e+01, -4.500000e+01, -5.500000e+01, -6.500000e+01, -7.500500e+01, -8.502500e+01, -9.509500e+01, -1.053100e+02, -1.158700e+02, -1.271500e+02, -1.397400e+02, -1.544700e+02, -1.724000e+02, -1.947350e+02, -2.227100e+02, -2.574700e+02, -2.999300e+02, -3.506800e+02, -4.099300e+02, -4.774700e+02, -5.527100e+02, -6.347350e+02, -7.224000e+02, -8.144700e+02, -9.097400e+02, -1.007155e+03, -1.105905e+03, -1.205535e+03, -1.306205e+03, -1.409150e+03, -1.517095e+03, -1.634175e+03, -1.765135e+03, -1.914150e+03, -2.084035e+03, -2.276225e+03, -2.491250e+03, -2.729250e+03, -2.990250e+03, -3.274250e+03, -3.581250e+03, -3.911250e+03, -4.264250e+03, -4.640250e+03, -5.039250e+03, -5.461250e+03, -5.906250e+03], dtype=float32)
- SSH(time, tile, j, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
array([[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.4194752 , -1.4261409 , -1.4262563 , ..., -1.335799 , -1.3286246 , -1.3223515 ], [-1.3915291 , -1.3982822 , -1.3976623 , ..., -1.3265072 , -1.3189714 , -1.3117282 ], [-1.3617151 , -1.3670161 , -1.3660299 , ..., -1.3197623 , -1.3118473 , -1.303166 ]], [[-1.3301036 , -1.3331454 , -1.3329679 , ..., -1.3132814 , -1.3051906 , -1.2948027 ], [-1.2997499 , -1.3010498 , -1.303525 , ..., -1.3048117 , -1.2970599 , -1.2852311 ], [-1.2769612 , -1.2781848 , -1.2847781 , ..., -1.2925622 , -1.2858012 , -1.2733294 ], ... [ 0.14350861, 0.14915593, 0.16067028, ..., -1.2250897 , -1.2384485 , -1.2555339 ], [ 0.14452162, 0.15083845, 0.1632475 , ..., -1.245257 , -1.2634263 , -1.2841328 ], [ 0.14675961, 0.15251625, 0.16487278, ..., -1.2580327 , -1.2814457 , -1.3074644 ]], [[-0.73443437, -0.8210122 , -0.907068 , ..., nan, nan, nan], [-0.7157703 , -0.8134533 , -0.9050429 , ..., nan, nan, nan], [-0.71003354, -0.81543124, -0.90919775, ..., nan, nan, nan], ..., [-1.2795744 , -1.3132012 , -1.3549699 , ..., nan, nan, nan], [-1.3077799 , -1.3369125 , -1.3719907 , ..., nan, nan, nan], [-1.3335364 , -1.3611741 , -1.3917223 , ..., nan, nan, nan]]]], dtype=float32) - ADVx_TH(time, k, tile, j, i_g)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Lateral advective flux of potential temperature in the model +x direction
- units :
- degree_C m3 s-1
- mate :
- ADVy_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +x direction through the 'u' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVx_TH(i_g,j,k) corresponds to +x fluxes through the 'u' face of the tracer cell at (i,j,k). Also, the model +x direction does not necessarily correspond to the geographical east-west direction because the x and y axes of the model's lat-lon-cap (llc) curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -28231902.0
- valid_max :
- 36523468.0
array([[[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ 1.19306547e+05, 1.09863953e+05, 1.08708008e+05, ..., 2.60564805e+04, 3.84371836e+04, 5.58860039e+04], [ 1.23932602e+05, 1.19640844e+05, 1.20704031e+05, ..., 3.50931055e+04, 5.25240781e+04, 6.95127891e+04], [ 1.29375188e+05, 1.30514531e+05, 1.32015797e+05, ..., 5.47346797e+04, 7.00478828e+04, 8.20443594e+04]], [[ 1.34879375e+05, 1.39183844e+05, 1.39052938e+05, ..., 7.06724141e+04, 7.57713438e+04, 6.89830312e+04], [ 1.31082719e+05, 1.34811516e+05, 1.30256555e+05, ..., 5.47624023e+04, 4.05062930e+04, 1.95837207e+04], [ 1.18207844e+05, 1.17642984e+05, 1.08316047e+05, ..., 4.20404961e+04, 2.74945742e+04, 2.43384043e+04], ... nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]]], dtype=float32) - ADVy_TH(time, k, tile, j_g, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Lateral advective flux of potential temperature in the model +y direction
- units :
- degree_C m3 s-1
- mate :
- ADVx_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +y direction through the 'v' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVy_TH(i,j_g,k) corresponds to +y fluxes through the 'v' face of the tracer cell at (i,j,k). Also, the model +y direction does not necessarily correspond to the geographical north-south direction because the x and y axes of the model's curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -31236064.0
- valid_max :
- 43466144.0
array([[[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-2.26423789e+04, -3.65008672e+04, -3.31573516e+04, ..., -3.80194766e+04, -3.46598438e+04, -3.05429355e+04], [-3.55484219e+04, -5.41644141e+04, -6.37171641e+04, ..., -2.48257168e+04, -1.77110176e+04, -8.63026172e+03], [-4.63738555e+04, -6.83228203e+04, -7.52612500e+04, ..., 2.51105234e+04, 2.88185957e+04, 2.93959648e+04]], [[-5.65837891e+04, -6.13085469e+04, -2.12589316e+04, ..., 3.19637090e+04, 3.32244141e+04, 3.06561016e+04], [ 3.63491133e+04, 4.60585430e+04, 7.47760781e+04, ..., 3.92454844e+04, 3.78334844e+04, 2.41646738e+04], [ 8.14258438e+04, 7.16582578e+04, 9.34466641e+04, ..., 4.91056484e+04, 3.51395039e+04, -2.40245781e+04], ... nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]]], dtype=float32)
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
1. Dimensions¶
Dimensions: (i: 90, j: 90, tile: 13, time: 12, i_g: 90, k: 50, j_g: 90)
ecco_dataset_merged is a container of DataArrays and as such it lists all of the unique dimensions its DataArrays. In other words, Dimensions shows all of the dimensions used by its variables.
2. Dimension Coordinates¶
Coordinates:
i (i) int32 0 1 2 3 4 5 6 7 8 9 … 80 81 82 83 84 85 86 87 88 89
j (j) int32 0 1 2 3 4 5 6 7 8 9 … 80 81 82 83 84 85 86 87 88 89
tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
time (time) datetime64[ns] 2010-01-16T12:00:00 … 2010-12-16T12:00:00
i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 … 80 81 82 83 84 85 86 87 88 89
k (k) int32 0 1 2 3 4 5 6 7 8 9 … 40 41 42 43 44 45 46 47 48 49
j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 … 80 81 82 83 84 85 86 87 88 89
Notice that the tile and time coordinates are unchanged. merge recognizes identical coordiantes and keeps them.
3. Non-Dimension Coordinates¶
XC (tile, j, i) float32 -111.60647 -111.303 -110.94285 … nan nan
YC (tile, j, i) float32 -88.24259 -88.382515 -88.52242 … nan nan
Z (k) float32 -5.0 -15.0 -25.0 -35.0 … -5039.25 -5461.25 -5906.25
The list of non-dimension coordinates includes the horizontal and vertical coordinates needed to locate each grid cell in geographical space. Like Dimensions, the non-dimension coordinates of the merged Dataset contain all of the non-dimension coordinates of the DataArrays. Notice that the horizontal coordinates of the grid corners (XG,YG) are not included, since they weren’t in any of the source DataArrays.
4. Attributes¶
Note that the high-level attributes of the new Dataset are just the attributes of the first DataArray in the merge, the SSH DataArray. The attributes of the Data variables remain intact:
[13]:
# (this json command makes Python dictionaries easier to read)
print(json.dumps(ecco_dataset_AB.ADVx_TH.attrs, indent=2,sort_keys=True))
{
"comment": "Lateral advective flux of potential temperature (THETA) in the +x direction through the 'u' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVx_TH(i_g,j,k) corresponds to +x fluxes through the 'u' face of the tracer cell at (i,j,k). Also, the model +x direction does not necessarily correspond to the geographical east-west direction because the x and y axes of the model's lat-lon-cap (llc) curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.",
"coverage_content_type": "modelResult",
"direction": ">0 increases potential temperature (THETA)",
"long_name": "Lateral advective flux of potential temperature in the model +x direction",
"mate": "ADVy_TH",
"units": "degree_C m3 s-1",
"valid_max": 36523468.0,
"valid_min": -28231902.0
}
Adding the model grid Dataset¶
Let’s use the merge routine to combine a Dataset of the model grid parameters with output_merged.
Load the model grid parameters¶
[14]:
# Load the llc90 grid parameters
import glob
grid_dataset = xr.open_dataset(glob.glob(join(ECCO_dir,'*GEOMETRY*','*.nc'))[0])
grid_dataset.coords
[14]:
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 10 ... 40 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* k_l (k_l) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* k_p1 (k_p1) int32 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49 50
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
XC (tile, j, i) float32 ...
YC (tile, j, i) float32 ...
XG (tile, j_g, i_g) float32 ...
YG (tile, j_g, i_g) float32 ...
Z (k) float32 ...
Zp1 (k_p1) float32 ...
Zu (k_u) float32 ...
Zl (k_l) float32 ...
XC_bnds (tile, j, i, nb) float32 ...
YC_bnds (tile, j, i, nb) float32 ...
Z_bnds (k, nv) float32 ...
Merge grid_all_tiles with output_merged¶
[15]:
ecco_dataset_ABG = xr.merge([ecco_dataset_AB, grid_dataset])
ecco_dataset_ABG
[15]:
<xarray.Dataset>
Dimensions: (i: 90, j: 90, tile: 13, time: 12, k: 50, i_g: 90, j_g: 90, k_u: 50, k_l: 50, k_p1: 51, nb: 4, nv: 2)
Coordinates: (12/21)
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
XC (tile, j, i) float32 -111.6 -111.3 -110.9 ... -99.42 -105.6 -111.9
YC (tile, j, i) float32 -88.24 -88.38 -88.52 ... -88.03 -88.08 -88.1
... ...
Zp1 (k_p1) float32 0.0 -10.0 -20.0 ... -5.244e+03 -5.678e+03 -6.134e+03
Zu (k_u) float32 -10.0 -20.0 -30.0 ... -5.678e+03 -6.134e+03
Zl (k_l) float32 0.0 -10.0 -20.0 ... -4.834e+03 -5.244e+03 -5.678e+03
XC_bnds (tile, j, i, nb) float32 ...
YC_bnds (tile, j, i, nb) float32 ...
Z_bnds (k, nv) float32 0.0 -10.0 -10.0 ... -5.678e+03 -6.134e+03
Dimensions without coordinates: nb, nv
Data variables: (12/24)
SSH (time, tile, j, i) float32 nan nan nan nan nan ... nan nan nan nan
ADVx_TH (time, k, tile, j, i_g) float32 nan nan nan nan ... nan nan nan nan
ADVy_TH (time, k, tile, j_g, i) float32 nan nan nan nan ... nan nan nan nan
CS (tile, j, i) float32 ...
SN (tile, j, i) float32 ...
rA (tile, j, i) float32 ...
... ...
hFacC (k, tile, j, i) float32 ...
hFacW (k, tile, j, i_g) float32 ...
hFacS (k, tile, j_g, i) float32 ...
maskC (k, tile, j, i) bool ...
maskW (k, tile, j, i_g) bool ...
maskS (k, tile, j_g, i) bool ...
Attributes:
long_name: Dynamic sea surface height anomaly
units: m
coverage_content_type: modelResult
standard_name: sea_surface_height_above_geoid
comment: Dynamic sea surface height anomaly above the geoi...
valid_min: -1.8805772066116333
valid_max: 1.4207719564437866- i: 90
- j: 90
- tile: 13
- time: 12
- k: 50
- i_g: 90
- j_g: 90
- k_u: 50
- k_l: 50
- k_p1: 51
- nb: 4
- nv: 2
- i(i)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at tracer and 'v' locations
- swap_dim :
- XC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'v' variables (e.g., VVEL) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j(j)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at tracer and 'u' locations
- swap_dim :
- YC
- comment :
- In the Arakawa C-grid system, tracer (e.g., THETA) and 'u' variables (e.g., UVEL) have the same y coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - tile(tile)int320 1 2 3 4 5 6 7 8 9 10 11 12
- long_name :
- lat-lon-cap tile index
- comment :
- The ECCO V4 horizontal model grid is divided into 13 tiles of 90x90 cells for convenience.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- time(time)datetime64[ns]2010-01-16T12:00:00 ... 2010-12-...
- long_name :
- center time of averaging period
- axis :
- T
- bounds :
- time_bnds
- coverage_content_type :
- coordinate
- standard_name :
- time
array(['2010-01-16T12:00:00.000000000', '2010-02-15T00:00:00.000000000', '2010-03-16T12:00:00.000000000', '2010-04-16T00:00:00.000000000', '2010-05-16T12:00:00.000000000', '2010-06-16T00:00:00.000000000', '2010-07-16T12:00:00.000000000', '2010-08-16T12:00:00.000000000', '2010-09-16T00:00:00.000000000', '2010-10-16T12:00:00.000000000', '2010-11-16T00:00:00.000000000', '2010-12-16T12:00:00.000000000'], dtype='datetime64[ns]') - XC(tile, j, i)float32-111.6 -111.3 ... -105.6 -111.9
- long_name :
- longitude of tracer grid cell center
- units :
- degrees_east
- coordinate :
- YC XC
- bounds :
- XC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-111.60647 , -111.303 , -110.94285 , ..., 64.791115, 64.80521 , 64.81917 ], [-104.8196 , -103.928444, -102.87706 , ..., 64.36745 , 64.41012 , 64.4524 ], [ -98.198784, -96.788055, -95.14185 , ..., 63.936497, 64.008224, 64.0793 ], ..., [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ]], [[ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], [ -37.5 , -36.5 , -35.5 , ..., 49.5 , 50.5 , 51.5 ], ... [ -40.5 , -40.5 , -40.5 , ..., -40.5 , -40.5 , -40.5 ], [ -39.5 , -39.5 , -39.5 , ..., -39.5 , -39.5 , -39.5 ], [ -38.5 , -38.5 , -38.5 , ..., -38.5 , -38.5 , -38.5 ]], [[-127.5 , -127.5 , -127.5 , ..., -115.850204, -115.50567 , -115.166985], [-126.5 , -126.5 , -126.5 , ..., -115.78025 , -115.464066, -115.153244], [-125.5 , -125.5 , -125.5 , ..., -115.71079 , -115.42275 , -115.139595], ..., [ -40.5 , -40.5 , -40.5 , ..., -101.42989 , -106.83081 , -112.28605 ], [ -39.5 , -39.5 , -39.5 , ..., -100.48844 , -106.24874 , -112.090065], [ -38.5 , -38.5 , -38.5 , ..., -99.42048 , -105.58465 , -111.86579 ]]], dtype=float32) - YC(tile, j, i)float32-88.24 -88.38 ... -88.08 -88.1
- long_name :
- latitude of tracer grid cell center
- units :
- degrees_north
- coordinate :
- YC XC
- bounds :
- YC_bnds
- comment :
- nonuniform grid spacing
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.24259 , -88.382515 , -88.52242 , ..., -80.57383 , -80.50492 , -80.43992 ], [-88.21676 , -88.354485 , -88.49178 , ..., -80.56925 , -80.50038 , -80.43542 ], [-88.164764 , -88.29827 , -88.43065 , ..., -80.55984 , -80.49105 , -80.42617 ], ..., [-58.321964 , -58.321964 , -58.321964 , ..., -58.321964 , -58.321964 , -58.321964 ], [-57.79962 , -57.79962 , -57.79962 , ..., -57.79962 , -57.79962 , -57.79962 ], [-57.271408 , -57.271408 , -57.271408 , ..., -57.271408 , -57.271408 , -57.271408 ]], [[-56.73891 , -56.73891 , -56.73891 , ..., -56.73891 , -56.73891 , -56.73891 ], [-56.2021 , -56.2021 , -56.2021 , ..., -56.2021 , -56.2021 , -56.2021 ], [-55.65936 , -55.65936 , -55.65936 , ..., -55.65936 , -55.65936 , -55.65936 ], ... [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ], [ 9.482398 , 8.516253 , 7.5699615, ..., -55.65936 , -56.2021 , -56.73891 ]], [[-57.271408 , -57.79962 , -58.321964 , ..., -80.36532 , -80.3745 , -80.37896 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.30862 , -80.317726 , -80.32216 ], [-57.271408 , -57.79962 , -58.321964 , ..., -80.25618 , -80.26523 , -80.26963 ], ..., [-57.271408 , -57.79962 , -58.321964 , ..., -87.7596 , -87.80198 , -87.822876 ], [-57.271408 , -57.79962 , -58.321964 , ..., -87.89526 , -87.94044 , -87.96276 ], [-57.271408 , -57.79962 , -58.321964 , ..., -88.030365 , -88.07871 , -88.10267 ]]], dtype=float32) - i_g(i_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- X
- long_name :
- grid index in x for variables at 'u' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- XG
- comment :
- In the Arakawa C-grid system, 'u' (e.g., UVEL) and 'g' variables (e.g., XG) have the same x coordinate on the model grid.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - j_g(j_g)int320 1 2 3 4 5 6 ... 84 85 86 87 88 89
- axis :
- Y
- long_name :
- grid index in y for variables at 'v' and 'g' locations
- c_grid_axis_shift :
- -0.5
- swap_dim :
- YG
- comment :
- In the Arakawa C-grid system, 'v' (e.g., VVEL) and 'g' variables (e.g., XG) have the same y coordinate.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89]) - k(k)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z for tracer variables
- swap_dim :
- Z
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - XG(tile, j_g, i_g)float32-115.0 -115.0 ... -102.9 -109.0
- long_name :
- longitude of 'southwest' corner of tracer grid cell
- units :
- degrees_east
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- longitude
array([[[-115. , -115. , -115. , ..., 65. , 65. , 65. ], [-108.47213 , -107.915245, -107.26045 , ..., 64.56598 , 64.59443 , 64.62261 ], [-101.9954 , -100.91797 , -99.66073 , ..., 64.12406 , 64.18147 , 64.23835 ], ..., [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ]], [[ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], [ -38. , -37. , -36. , ..., 49. , 50. , 51. ], ... [ -41. , -41. , -41. , ..., -41. , -41. , -41. ], [ -40. , -40. , -40. , ..., -40. , -40. , -40. ], [ -39. , -39. , -39. , ..., -39. , -39. , -39. ]], [[-128. , -128. , -128. , ..., -116.06686 , -115.70526 , -115.34945 ], [-127. , -127. , -127. , ..., -115.982254, -115.64933 , -115.32173 ], [-126. , -126. , -126. , ..., -115.89828 , -115.59381 , -115.29422 ], ..., [ -41. , -41. , -41. , ..., -99.28857 , -104.466286, -109.73619 ], [ -40. , -40. , -40. , ..., -98.24634 , -103.74563 , -109.36924 ], [ -39. , -39. , -39. , ..., -97.07265 , -102.928925, -108.95171 ]]], dtype=float32) - YG(tile, j_g, i_g)float32-88.18 -88.32 ... -87.99 -88.02
- long_name :
- latitude of 'southwest' corner of tracer grid cell
- units :
- degrees_north
- coordinate :
- YG XG
- comment :
- Nonuniform grid spacing. Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- standard_name :
- latitude
array([[[-88.17569 , -88.31587 , -88.45609 , ..., -80.610275, -80.53945 , -80.47247 ], [-88.16335 , -88.30251 , -88.44153 , ..., -80.608 , -80.5372 , -80.470245], [-88.12586 , -88.26202 , -88.397514, ..., -80.601006, -80.530266, -80.46337 ], ..., [-58.58002 , -58.58002 , -58.58002 , ..., -58.58002 , -58.58002 , -58.58002 ], [-58.06183 , -58.06183 , -58.06183 , ..., -58.06183 , -58.06183 , -58.06183 ], [-57.53605 , -57.53605 , -57.53605 , ..., -57.53605 , -57.53605 , -57.53605 ]], [[-57.005695, -57.005695, -57.005695, ..., -57.005695, -57.005695, -57.005695], [-56.471046, -56.471046, -56.471046, ..., -56.471046, -56.471046, -56.471046], [-55.931786, -55.931786, -55.931786, ..., -55.931786, -55.931786, -55.931786], ... [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046], [ 9.96973 , 8.997536, 8.039881, ..., -55.384766, -55.931786, -56.471046]], [[-57.005695, -57.53605 , -58.06183 , ..., -80.38878 , -80.400444, -80.40726 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.33003 , -80.34161 , -80.34837 ], [-57.005695, -57.53605 , -58.06183 , ..., -80.27549 , -80.286995, -80.29371 ], ..., [-57.005695, -57.53605 , -58.06183 , ..., -87.66318 , -87.71477 , -87.7454 ], [-57.005695, -57.53605 , -58.06183 , ..., -87.797325, -87.85214 , -87.884766], [-57.005695, -57.53605 , -58.06183 , ..., -87.93075 , -87.9892 , -88.02409 ]]], dtype=float32) - Z(k)float32-5.0 -15.0 ... -5.906e+03
- long_name :
- depth of tracer grid cell center
- units :
- m
- positive :
- up
- bounds :
- Z_bnds
- comment :
- Non-uniform vertical spacing.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([-5.000000e+00, -1.500000e+01, -2.500000e+01, -3.500000e+01, -4.500000e+01, -5.500000e+01, -6.500000e+01, -7.500500e+01, -8.502500e+01, -9.509500e+01, -1.053100e+02, -1.158700e+02, -1.271500e+02, -1.397400e+02, -1.544700e+02, -1.724000e+02, -1.947350e+02, -2.227100e+02, -2.574700e+02, -2.999300e+02, -3.506800e+02, -4.099300e+02, -4.774700e+02, -5.527100e+02, -6.347350e+02, -7.224000e+02, -8.144700e+02, -9.097400e+02, -1.007155e+03, -1.105905e+03, -1.205535e+03, -1.306205e+03, -1.409150e+03, -1.517095e+03, -1.634175e+03, -1.765135e+03, -1.914150e+03, -2.084035e+03, -2.276225e+03, -2.491250e+03, -2.729250e+03, -2.990250e+03, -3.274250e+03, -3.581250e+03, -3.911250e+03, -4.264250e+03, -4.640250e+03, -5.039250e+03, -5.461250e+03, -5.906250e+03], dtype=float32) - k_u(k_u)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z corresponding to the bottom face of tracer grid cells ('w' locations)
- c_grid_axis_shift :
- 0.5
- swap_dim :
- Zu
- comment :
- First index corresponds to the bottom face of the uppermost tracer grid cell. The use of 'u' in the variable name follows the MITgcm convention for naming the bottom face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_l(k_l)int320 1 2 3 4 5 6 ... 44 45 46 47 48 49
- axis :
- Z
- long_name :
- grid index in z corresponding to the top face of tracer grid cells ('w' locations)
- c_grid_axis_shift :
- -0.5
- swap_dim :
- Zl
- comment :
- First index corresponds to the top face of the uppermost tracer grid cell. The use of 'l' in the variable name follows the MITgcm convention for naming the top face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - k_p1(k_p1)int320 1 2 3 4 5 6 ... 45 46 47 48 49 50
- axis :
- Z
- long_name :
- grid index in z for variables at 'w' locations
- c_grid_axis_shift :
- [-0.5 0.5]
- swap_dim :
- Zp1
- comment :
- Includes top of uppermost model tracer cell (k_p1=0) and bottom of lowermost tracer cell (k_p1=51).
- coverage_content_type :
- coordinate
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]) - Zp1(k_p1)float32...
- long_name :
- depth of top/bottom face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- Contains one element more than the number of vertical layers. First element is 0m, the depth of the top face of the uppermost grid cell. Last element is the depth of the bottom face of the deepest grid cell.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zu(k_u)float32...
- long_name :
- depth of bottom face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- First element is -10m, the depth of the bottom face of the uppermost tracer grid cell. Last element is the depth of the bottom face of the deepest grid cell. The use of 'u' in the variable name follows the MITgcm convention for naming the bottom face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. , -6134.5 ], dtype=float32) - Zl(k_l)float32...
- long_name :
- depth of top face of tracer grid cell
- units :
- m
- positive :
- up
- comment :
- First element is 0m, the depth of the top face of the uppermost tracer grid cell (i.e., the ocean surface). Last element is the depth of the top face of the deepest grid cell. The use of 'l' in the variable name follows the MITgcm convention for naming the top face of ocean tracer grid cells.
- coverage_content_type :
- coordinate
- standard_name :
- depth
array([ 0. , -10. , -20. , -30. , -40. , -50. , -60. , -70. , -80.01, -90.04, -100.15, -110.47, -121.27, -133.03, -146.45, -162.49, -182.31, -207.16, -238.26, -276.68, -323.18, -378.18, -441.68, -513.26, -592.16, -677.31, -767.49, -861.45, -958.03, -1056.28, -1155.53, -1255.54, -1356.87, -1461.43, -1572.76, -1695.59, -1834.68, -1993.62, -2174.45, -2378. , -2604.5 , -2854. , -3126.5 , -3422. , -3740.5 , -4082. , -4446.5 , -4834. , -5244.5 , -5678. ], dtype=float32) - XC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. XC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), XC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), XC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), XC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- longitudes of tracer grid cell corners
[421200 values with dtype=float32]
- YC_bnds(tile, j, i, nb)float32...
- comment :
- Bounds array follows CF conventions. YC_bnds[i,j,0] = 'southwest' corner (j-1, i-1), YC_bnds[i,j,1] = 'southeast' corner (j-1, i+1), YC_bnds[i,j,2] = 'northeast' corner (j+1, i+1), YC_bnds[i,j,3] = 'northwest' corner (j+1, i-1). Note: 'southwest', 'southeast', northwest', and 'northeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm dcoumentation for details.
- coverage_content_type :
- coordinate
- long_name :
- latitudes of tracer grid cell corners
[421200 values with dtype=float32]
- Z_bnds(k, nv)float32...
- comment :
- One pair of depths for each vertical level.
- coverage_content_type :
- coordinate
- long_name :
- depths of top and bottom faces of tracer grid cell
array([[ 0. , -10. ], [ -10. , -20. ], [ -20. , -30. ], [ -30. , -40. ], [ -40. , -50. ], [ -50. , -60. ], [ -60. , -70. ], [ -70. , -80.01 ], [ -80.01 , -90.04 ], [ -90.04 , -100.15 ], [ -100.15 , -110.47 ], [ -110.47 , -121.270004], [ -121.270004, -133.03 ], [ -133.03 , -146.45 ], [ -146.45 , -162.48999 ], [ -162.48999 , -182.31 ], [ -182.31 , -207.16 ], [ -207.16 , -238.26001 ], [ -238.26001 , -276.68 ], [ -276.68 , -323.18 ], [ -323.18 , -378.18 ], [ -378.18 , -441.68 ], [ -441.68 , -513.26 ], [ -513.26 , -592.16003 ], [ -592.16003 , -677.31006 ], [ -677.31006 , -767.49005 ], [ -767.49005 , -861.4501 ], [ -861.4501 , -958.0301 ], [ -958.0301 , -1056.28 ], [-1056.28 , -1155.53 ], [-1155.53 , -1255.54 ], [-1255.54 , -1356.87 ], [-1356.87 , -1461.4299 ], [-1461.4299 , -1572.7599 ], [-1572.7599 , -1695.5898 ], [-1695.5898 , -1834.6798 ], [-1834.6798 , -1993.6199 ], [-1993.6199 , -2174.45 ], [-2174.45 , -2378. ], [-2378. , -2604.5 ], [-2604.5 , -2854. ], [-2854. , -3126.5 ], [-3126.5 , -3422. ], [-3422. , -3740.5 ], [-3740.5 , -4082. ], [-4082. , -4446.5 ], [-4446.5 , -4834. ], [-4834. , -5244.5 ], [-5244.5 , -5678. ], [-5678. , -6134.5 ]], dtype=float32)
- SSH(time, tile, j, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
array([[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.4194752 , -1.4261409 , -1.4262563 , ..., -1.335799 , -1.3286246 , -1.3223515 ], [-1.3915291 , -1.3982822 , -1.3976623 , ..., -1.3265072 , -1.3189714 , -1.3117282 ], [-1.3617151 , -1.3670161 , -1.3660299 , ..., -1.3197623 , -1.3118473 , -1.303166 ]], [[-1.3301036 , -1.3331454 , -1.3329679 , ..., -1.3132814 , -1.3051906 , -1.2948027 ], [-1.2997499 , -1.3010498 , -1.303525 , ..., -1.3048117 , -1.2970599 , -1.2852311 ], [-1.2769612 , -1.2781848 , -1.2847781 , ..., -1.2925622 , -1.2858012 , -1.2733294 ], ... [ 0.14350861, 0.14915593, 0.16067028, ..., -1.2250897 , -1.2384485 , -1.2555339 ], [ 0.14452162, 0.15083845, 0.1632475 , ..., -1.245257 , -1.2634263 , -1.2841328 ], [ 0.14675961, 0.15251625, 0.16487278, ..., -1.2580327 , -1.2814457 , -1.3074644 ]], [[-0.73443437, -0.8210122 , -0.907068 , ..., nan, nan, nan], [-0.7157703 , -0.8134533 , -0.9050429 , ..., nan, nan, nan], [-0.71003354, -0.81543124, -0.90919775, ..., nan, nan, nan], ..., [-1.2795744 , -1.3132012 , -1.3549699 , ..., nan, nan, nan], [-1.3077799 , -1.3369125 , -1.3719907 , ..., nan, nan, nan], [-1.3335364 , -1.3611741 , -1.3917223 , ..., nan, nan, nan]]]], dtype=float32) - ADVx_TH(time, k, tile, j, i_g)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Lateral advective flux of potential temperature in the model +x direction
- units :
- degree_C m3 s-1
- mate :
- ADVy_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +x direction through the 'u' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVx_TH(i_g,j,k) corresponds to +x fluxes through the 'u' face of the tracer cell at (i,j,k). Also, the model +x direction does not necessarily correspond to the geographical east-west direction because the x and y axes of the model's lat-lon-cap (llc) curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -28231902.0
- valid_max :
- 36523468.0
array([[[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ 1.19306547e+05, 1.09863953e+05, 1.08708008e+05, ..., 2.60564805e+04, 3.84371836e+04, 5.58860039e+04], [ 1.23932602e+05, 1.19640844e+05, 1.20704031e+05, ..., 3.50931055e+04, 5.25240781e+04, 6.95127891e+04], [ 1.29375188e+05, 1.30514531e+05, 1.32015797e+05, ..., 5.47346797e+04, 7.00478828e+04, 8.20443594e+04]], [[ 1.34879375e+05, 1.39183844e+05, 1.39052938e+05, ..., 7.06724141e+04, 7.57713438e+04, 6.89830312e+04], [ 1.31082719e+05, 1.34811516e+05, 1.30256555e+05, ..., 5.47624023e+04, 4.05062930e+04, 1.95837207e+04], [ 1.18207844e+05, 1.17642984e+05, 1.08316047e+05, ..., 4.20404961e+04, 2.74945742e+04, 2.43384043e+04], ... nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]]], dtype=float32) - ADVy_TH(time, k, tile, j_g, i)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Lateral advective flux of potential temperature in the model +y direction
- units :
- degree_C m3 s-1
- mate :
- ADVx_TH
- coverage_content_type :
- modelResult
- direction :
- >0 increases potential temperature (THETA)
- comment :
- Lateral advective flux of potential temperature (THETA) in the +y direction through the 'v' face of the tracer cell on the native model grid. Note: in the Arakawa-C grid, horizontal flux quantities are staggered relative to the tracer cells with indexing such that +ADVy_TH(i,j_g,k) corresponds to +y fluxes through the 'v' face of the tracer cell at (i,j,k). Also, the model +y direction does not necessarily correspond to the geographical north-south direction because the x and y axes of the model's curvilinear lat-lon-cap (llc) grid have arbitrary orientations which vary within and across tiles.
- valid_min :
- -31236064.0
- valid_max :
- 43466144.0
array([[[[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-2.26423789e+04, -3.65008672e+04, -3.31573516e+04, ..., -3.80194766e+04, -3.46598438e+04, -3.05429355e+04], [-3.55484219e+04, -5.41644141e+04, -6.37171641e+04, ..., -2.48257168e+04, -1.77110176e+04, -8.63026172e+03], [-4.63738555e+04, -6.83228203e+04, -7.52612500e+04, ..., 2.51105234e+04, 2.88185957e+04, 2.93959648e+04]], [[-5.65837891e+04, -6.13085469e+04, -2.12589316e+04, ..., 3.19637090e+04, 3.32244141e+04, 3.06561016e+04], [ 3.63491133e+04, 4.60585430e+04, 7.47760781e+04, ..., 3.92454844e+04, 3.78334844e+04, 2.41646738e+04], [ 8.14258438e+04, 7.16582578e+04, 9.34466641e+04, ..., 4.91056484e+04, 3.51395039e+04, -2.40245781e+04], ... nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]]], dtype=float32) - CS(tile, j, i)float32...
- long_name :
- cosine of tracer grid cell orientation vs geographical north
- units :
- 1
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- comment :
- CS and SN are required to calculate the geographic (meridional, zonal) components of vectors on the curvilinear model grid. Note: for vector R with components R_x and R_y: R_{east} = CS R_x - SN R_y. R_{north} = SN R_x + CS R_y
[105300 values with dtype=float32]
- SN(tile, j, i)float32...
- long_name :
- sine of tracer grid cell orientation vs geographical north
- units :
- 1
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- comment :
- CS and SN are required to calculate the geographic (meridional, zonal) components of vectors on the curvilinear model grid. Note: for vector R with components R_x and R_y in local grid directions x and y, the geographical eastward component R_{east} = CS R_x - SN R_y. The geographical northward component R_{north} = SN R_x + CS R_y.
[105300 values with dtype=float32]
- rA(tile, j, i)float32...
- long_name :
- area of tracer grid cell
- units :
- m2
- coordinate :
- YC XC
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
[105300 values with dtype=float32]
- dxG(tile, j_g, i)float32...
- long_name :
- distance between 'southwest' and 'southeast' corners of the tracer grid cell
- units :
- m
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'south' side of tracer grid cell. Note: 'south', 'southwest', and 'southeast' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- dyG(tile, j, i_g)float32...
- long_name :
- distance between 'southwest' and 'northwest' corners of the tracer grid cell
- units :
- m
- coordinate :
- YC XG
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'west' side of tracer grid cell. Note: 'west, 'southwest', and 'northwest' do not correspond to geographic orientation but are used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- Depth(tile, j, i)float32...
- long_name :
- model seafloor depth below ocean surface at rest
- units :
- m
- coordinate :
- XC YC
- coverage_content_type :
- modelResult
- standard_name :
- sea_floor_depth_below_geoid
- comment :
- Model sea surface height (SSH) of 0m corresponds to an ocean surface at rest relative to the geoid. Depth corresponds to seafloor depth below geoid. Note: the MITgcm used by ECCO V4r4 implements 'partial cells' so the actual model seafloor depth may differ from the seafloor depth provided by the input bathymetry file.
[105300 values with dtype=float32]
- rAz(tile, j_g, i_g)float32...
- long_name :
- area of vorticity 'g' grid cell
- units :
- m2
- coordinate :
- YG XG
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Vorticity cells are staggered in space relative to tracer cells, nominally situated on tracer cell corners. Vorticity cell (i,j) is located at the 'southwest' corner of tracer grid cell (i, j). Note: 'southwest' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- dxC(tile, j, i_g)float32...
- long_name :
- distance between centers of adjacent tracer grid cells in the 'x' direction
- units :
- m
- coordinate :
- YC XG
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'north' side of vorticity grid cells. Note: 'north' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- dyC(tile, j_g, i)float32...
- long_name :
- distance between centers of adjacent tracer grid cells in the 'y' direction
- units :
- m
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- comment :
- Alternatively, the length of 'east' side of vorticity grid cells. Note: 'east' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- rAw(tile, j, i_g)float32...
- long_name :
- area of 'v' grid cell
- units :
- m2
- coordinate :
- YG XC
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Model 'v' grid cells are staggered in space between adjacent tracer grid cells in the 'x' direction. 'v' grid cell (i,j) is situated at the 'west' edge of tracer grid cell (i, j). Note: 'west' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- rAs(tile, j_g, i)float32...
- long_name :
- area of 'u' grid cell
- units :
- m2
- coverage_content_type :
- modelResult
- standard_name :
- cell_area
- comment :
- Model 'u' grid cells are staggered in space between adjacent tracer grid cells in the 'y' direction. 'u' grid cell (i,j) is situated at the 'south' edge of tracer grid cell (i, j). Note: 'south' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[105300 values with dtype=float32]
- drC(k_p1)float32...
- long_name :
- distance between the centers of adjacent tracer grid cells in the 'z' direction
- units :
- m
- coverage_content_type :
- modelResult
- comment :
- The first element corresponds to the distance between the depth of the center of the uppermost model grid cell and the surface.
array([ 5. , 10. , 10. , 10. , 10. , 10. , 10. , 10.005, 10.02 , 10.07 , 10.215, 10.56 , 11.28 , 12.59 , 14.73 , 17.93 , 22.335, 27.975, 34.76 , 42.46 , 50.75 , 59.25 , 67.54 , 75.24 , 82.025, 87.665, 92.07 , 95.27 , 97.415, 98.75 , 99.63 , 100.67 , 102.945, 107.945, 117.08 , 130.96 , 149.015, 169.885, 192.19 , 215.025, 238. , 261. , 284. , 307. , 330. , 353. , 376. , 399. , 422. , 445. , 228.25 ], dtype=float32) - drF(k)float32...
- long_name :
- distance between the upper and lower interfaces of the model grid cell
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- cell_thickness
- comment :
- Nominal grid cell thickness. Note: in the z* coordinate system used in ECCO V4, actual tracer grid cell thickness, h, varies through time as h(i,j,k,t)= drF(k) hfacC(i,j,k,t).
array([ 10. , 10. , 10. , 10. , 10. , 10. , 10. , 10.01, 10.03, 10.11, 10.32, 10.8 , 11.76, 13.42, 16.04, 19.82, 24.85, 31.1 , 38.42, 46.5 , 55. , 63.5 , 71.58, 78.9 , 85.15, 90.18, 93.96, 96.58, 98.25, 99.25, 100.01, 101.33, 104.56, 111.33, 122.83, 139.09, 158.94, 180.83, 203.55, 226.5 , 249.5 , 272.5 , 295.5 , 318.5 , 341.5 , 364.5 , 387.5 , 410.5 , 433.5 , 456.5 ], dtype=float32) - PHrefC(k)float32...
- long_name :
- reference ocean hydrostatic pressure at tracer grid cell center
- units :
- m2 s-2
- coverage_content_type :
- modelResult
- comment :
- PHrefC = p_ref (k) / rhoConst = rhoConst g z(k) / rhoConst = g z(k), where p_ref(k) is reference hydrostatic ocean pressure at center of tracer grid cell k, rhoConst is reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and z(k) is depth at center of tracer grid cell k. Units: p:[kg m-1 s-2], rhoConst:[kg m-3], g:[m s-2], z_m(t):[m]. Note: does not include atmospheric pressure loading. Quantity referred to in some contexts as hydrostatic pressure potential. PHIHYDcR is anomaly of PHrefC.
array([4.905000e+01, 1.471500e+02, 2.452500e+02, 3.433500e+02, 4.414500e+02, 5.395500e+02, 6.376500e+02, 7.357991e+02, 8.340953e+02, 9.328820e+02, 1.033091e+03, 1.136685e+03, 1.247342e+03, 1.370849e+03, 1.515351e+03, 1.691244e+03, 1.910350e+03, 2.184785e+03, 2.525781e+03, 2.942313e+03, 3.440171e+03, 4.021413e+03, 4.683980e+03, 5.422085e+03, 6.226750e+03, 7.086744e+03, 7.989951e+03, 8.924550e+03, 9.880190e+03, 1.084893e+04, 1.182630e+04, 1.281387e+04, 1.382376e+04, 1.488270e+04, 1.603126e+04, 1.731597e+04, 1.877781e+04, 2.044438e+04, 2.232977e+04, 2.443916e+04, 2.677394e+04, 2.933435e+04, 3.212039e+04, 3.513206e+04, 3.836936e+04, 4.183229e+04, 4.552085e+04, 4.943504e+04, 5.357486e+04, 5.794031e+04], dtype=float32) - PHrefF(k_p1)float32...
- long_name :
- reference ocean hydrostatic pressure at tracer grid cell top/bottom interface
- units :
- m2 s-2
- coverage_content_type :
- modelResult
- comment :
- PHrefF = p_ref (k_l) / rhoConst = rhoConst g z(k_l) / rhoConst = g z(k_l), where p_ref(k_l) is reference hydrostatic ocean pressure at lower interface of tracer grid cell k, rhoConst is reference density (1029 kg m-3), g is acceleration due to gravity (9.81 m s-2), and z(k) is depth at center of tracer grid cell k. Units: p:[kg m-1 s-2], rhoConst:[kg m-3], g:[m s-2], z_m(t):[m]. Note: does not include atmospheric pressure loading. Quantity referred to in some contexts as hydrostatic pressure potential. See PHrefC
array([ 0. , 98.1 , 196.2 , 294.3 , 392.4 , 490.5 , 588.6 , 686.7 , 784.8981, 883.2924, 982.4715, 1083.7107, 1189.6587, 1305.0243, 1436.6746, 1594.0269, 1788.461 , 2032.2396, 2337.3306, 2714.2307, 3170.3958, 3709.9458, 4332.881 , 5035.0806, 5809.09 , 6644.411 , 7529.0767, 8450.824 , 9398.274 , 10362.106 , 11335.749 , 12316.848 , 13310.895 , 14336.628 , 15428.775 , 16633.738 , 17998.21 , 19557.412 , 21331.355 , 23328.18 , 25550.145 , 27997.74 , 30670.965 , 33569.82 , 36694.305 , 40044.42 , 43620.164 , 47421.54 , 51448.547 , 55701.18 , 60179.445 ], dtype=float32) - hFacC(k, tile, j, i)float32...
- long_name :
- vertical open fraction of tracer grid cell
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- Tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacC. The model allows for partially-filled cells to represent topographic variations more smoothly (hFacC < 1). Completely closed (dry) tracer grid cells have hFacC = 0. Note: the model z* coordinate system allows hFacC to vary through time. A time-invariant hFacC field is provided for reference.
[5265000 values with dtype=float32]
- hFacW(k, tile, j, i_g)float32...
- long_name :
- vertical open fraction of tracer grid cell 'west' face
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- The 'west' face of tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacW. The model allows for partially-filled cells for smoother representation of seafloor topography. Tracer grid cells adjacent in the 'x' direction that are partially closed in the vertical have hFacW < 1. The model z* coordinate system used by the model permits hFacC, and therefore hFacW, to vary through time. A time-invariant hFacW field is provided for reference. Note: The term 'west' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[5265000 values with dtype=float32]
- hFacS(k, tile, j_g, i)float32...
- long_name :
- vertical open fraction of tracer grid cell 'south' face
- coverage_content_type :
- modelResult
- units :
- 1
- comment :
- The 'south' face of tracer grid cells may be fractionally closed in the vertical. The open vertical fraction is hFacS. The model allows for partially-filled cells for smoother representation of seafloor topography. Tracer grid cells adjacent in the 'y' direction that are partially closed in the vertical have hFacS < 1. The model z* coordinate system used by the model permits hFacC, and therefore hFacS, to vary through time. A time-invariant hFacS field is provided for reference. Note: The term 'south' does not correspond to geographic orientation but is used for convenience to describe the computational grid. See MITgcm documentation for details.
[5265000 values with dtype=float32]
- maskC(k, tile, j, i)bool...
- long_name :
- wet/dry boolean mask for tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for tracer grid cells with nonzero open vertical fraction (hFacC > 0), otherwise False. Although hFacC can vary though time, cells will never close if starting open and will never open if starting closed: hFacC(i,j,k,t) > 0 for all t, if hFacC(i,j,k,t=0) and hFacC(i,j,k,t) = 0 for all t, if hFacC(i,j,k,t=0) = 0. Therefore, maskC is time invariant.
[5265000 values with dtype=bool]
- maskW(k, tile, j, i_g)bool...
- long_name :
- wet/dry boolean mask for 'west' face of tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for grid cells with nonzero open vertical fraction along their 'west' face (hFacW > 0), otherwise False. Although hFacW can vary though time, cells will never close if starting open and will never open if starting closed: hFacW(i,j,k,t) > 0 for all t, if hFacW(i,j,k,t=0) and hFacW(i,j,k,t) = 0 for all t, if hFacW(i,j,k,t=0) = 0. Therefore, maskW is time invariant. Note:
[5265000 values with dtype=bool]
- maskS(k, tile, j_g, i)bool...
- long_name :
- wet/dry boolean mask for 'south' face of tracer grid cell
- coverage_content_type :
- modelResult
- comment :
- True for grid cells with nonzero open vertical fraction along their 'south' face (hFacS > 0), otherwise False. Although hFacS can vary though time, cells will never close if starting open and will never open if starting closed: hFacS(i,j,k,t) > 0 for all t, if hFacS(i,j,k,t=0) and hFacS(i,j,k,t) = 0 for all t, if hFacS(i,j,k,t=0) = 0. Therefore, maskS is time invariant. Note:
[5265000 values with dtype=bool]
- long_name :
- Dynamic sea surface height anomaly
- units :
- m
- coverage_content_type :
- modelResult
- standard_name :
- sea_surface_height_above_geoid
- comment :
- Dynamic sea surface height anomaly above the geoid, suitable for comparisons with altimetry sea surface height data products that apply the inverse barometer (IB) correction. Note: SSH is calculated by correcting model sea level anomaly ETAN for three effects: a) global mean steric sea level changes related to density changes in the Boussinesq volume-conserving model (Greatbatch correction, see sterGloH), b) the inverted barometer (IB) effect (see SSHIBC) and c) sea level displacement due to sea-ice and snow pressure loading (see sIceLoad). SSH can be compared with the similarly-named SSH variable in previous ECCO products that did not include atmospheric pressure loading (e.g., Version 4 Release 3). Use SSHNOIBC for comparisons with altimetry data products that do NOT apply the IB correction.
- valid_min :
- -1.8805772066116333
- valid_max :
- 1.4207719564437866
Examining the merged Dataset¶
The result of this last merge is a single Dataset with 3 Data variables, and a complete set of model grid parameters (geographical coordinates, distances, areas), including the coordinates of the grid cell corners XG,YG.
Merging and memory¶
Merging Datasets together does not make copies of the data in memory. Instead, merged Datasets are in fact just a reorganized collection of pointers. You may want to delete the original variables to clear your namespace, but it is not necessary.
Summary¶
Now you know how to merge multiple Datasets using the merge command. We demonstrated merging of Datasets constructed from three different variables types and the model grid parameters.