The Dataset and DataArray objects used in the ECCOv4 Python package.¶
Objectives¶
To introduce the two high-level data structures, Dataset and DataArray, that are used in by the ecco_v4_py Python package to load and store the ECCO v4 model grid parameters and state estimate variables.
Introduction¶
The ECCO version 4 release 4 (v4r4) files are provided as NetCDF files. This tutorial shows you how to download and open these files using Python code, and takes a look at the structure of these files. The ECCO output is available as a number of datasets that each contain a few variables. Each dataset consists of files corresponding to a single time coordinate (monthly mean, daily mean, or snapshot). Each dataset file that represents a single time is called a granule.
In this first tutorial we will start slowly, providing detail at every step. Later tutorials will assume knowledge of some basic operations introduced here.
Let’s get started.
Import external packages and modules¶
Before using Python libraries we must import them. Usually this is done at the beginning of every Python program or interactive Juypter notebook instance but one can import a library at any point in the code. Python libraries, called packages, contain subroutines and/or define data structures that provide useful functionality.
Before we go further, let’s import some packages needed for this tutorial:
[1]:
# NumPy is the fundamental package for scientific computing with Python.
# It contains among other things:
# a powerful N-dimensional array object
# sophisticated (broadcasting) functions
# tools for integrating C/C++ and Fortran code
# useful linear algebra, Fourier transform, and random number capabilities
# http://www.numpy.org/
#
# make all functions from the 'numpy' module available with the prefix 'np'
import numpy as np
# xarray is an open source project and Python package that aims to bring the
# labeled data power of pandas to the physical sciences, by providing
# N-dimensional variants of the core pandas data structures.
# Our approach adopts the Common Data Model for self- describing scientific
# data in widespread use in the Earth sciences: xarray.Dataset is an in-memory
# representation of a netCDF file.
# http://xarray.pydata.org/en/stable/
#
# import all function from the 'xarray' module available with the prefix 'xr'
import xarray as xr
Load the ECCO Version 4 Python package¶
The ecco_v4_py is a Python package written specifically for working with ECCO NetCDF output.
See the “Getting Started” page in the tutorial for instructions about installing the ecco_v4_py module on your machine.
[2]:
## Import the ecco_v4_py library into Python
## =========================================
## -- If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it using sys.path.append.
## For example, if your ecco_v4_py files are in ~/ECCOv4-py/ecco_v4_py,
## you can use:
from os.path import expanduser,join,isdir
import sys
user_home_dir = expanduser('~')
ecco_v4_py_dir = join(user_home_dir,'ECCOv4-py')
if isdir(ecco_v4_py_dir):
sys.path.append(ecco_v4_py_dir)
import ecco_v4_py as ecco
The syntax
import XYZ package as ABC
allows you to access all of the subroutines and/or objects in a package with perhaps a long complicated name with a shorter, easier name.
Here, we import ecco_v4_py as ecco because typing ecco is easier than ecco_v4_py every time. Also, ecco_v4_py is actually comprised of multiple python modules and by importing just ecco_v4_py we can actually access all of the subroutines in those modules as well. Fancy.
Downloading and opening state estimate NetCDF files (datasets)¶
You can access the ECCOv4r4 files through PO.DAAC, either by downloading them to your own machine, or downloading or opening them from an AWS Cloud instance. The ecco_download.py module helps with downloads to your own machine, while the ecco_s3_retrieve.py module will help
download or access files when you are working in a cloud instance. To indicate that you are working in an instance, specify incloud_access = True, otherwise it is assumed you are working outside of the cloud. Directories can be appended to your path using sys.path.append.
To open ECCO v4’s NetCDF files we will use the open_mfdataset command from the Python package xarray. xarray has the open_dataset routine which creates a Dataset object and loads the contents of the NetCDF file, including its metadata, into a data structure. The open_mfdataset routine does the same thing, but also concatenates multiple netCDF files with compatible dimensions and coordinates–a very handy feature!
Let’s download and open the monthly mean temperature/salinity files for 2010.
[3]:
# indicate whether you are working in a cloud instance (True if yes, False otherwise)
incloud_access = False
# ECCO_dir specifies parent directory of all ECCOv4r4 downloads
# ECCO_dir = None downloads to default path ~/Downloads/ECCO_V4r4_PODAAC/
ECCO_dir = None
ShortNames_list = ["ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
if incloud_access == True:
from ecco_s3_retrieve import ecco_podaac_s3_get_diskaware
# retrieve files (download to instance if there is sufficient storage)
files_nested_list = ecco_podaac_s3_get_diskaware(ShortNames=ShortNames_list,\
StartDate='2010-01',EndDate='2010-12',\
max_avail_frac=0.5,\
download_root_dir=ECCO_dir)
# load file into workspace
ds = xr.open_mfdataset(files_nested_list[ShortNames_list[0]],compat='override',data_vars='minimal',coords='minimal')
else:
# load module into workspace (if module is not in current directory you should add it to your path using sys.path.append)
from ecco_download import ecco_podaac_download
# download files (granules) containing 2010 monthly mean temperatures
files_list = ecco_podaac_download(ShortName=ShortNames_list[0],\
StartDate="2010-01",EndDate="2010-12",\
download_root_dir=ECCO_dir,n_workers=6,force_redownload=False)
ds = xr.open_mfdataset(files_list,compat='override',data_vars='minimal',coords='minimal')
created download directory C:\Users\adelman\Downloads\ECCO_V4r4_PODAAC\ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4
Total number of matching granules: 12
DL Progress: 100%|#########################| 12/12 [00:15<00:00, 1.29s/it]
=====================================
total downloaded: 208.75 Mb
avg download speed: 13.48 Mb/s
Time spent = 15.480218887329102 seconds
What is ds? It is a Dataset object which is defined somewhere deep in the xarray package:
[4]:
type(ds)
[4]:
xarray.core.dataset.Dataset
The Dataset object¶
According to the xarray documentation, a Dataset is a Python object designed as an “in-memory representation of the data model from the NetCDF file format.”
What does that mean? NetCDF files are self-describing in the sense that they include information about the data they contain. When Datasets are created by loading a NetCDF file they load all of the same data and metadata.
Just as a NetCDF file can contain many variables, a Dataset can contain many variables. These variables are referred to as Data Variables in the xarray nomenclature.
Datasets contain three main classes of fields:
Coordinates : arrays identifying the coordinates of the data variables
Data Variables: the data variable arrays and their associated coordinates
Attributes : metadata describing the dataset
Now that we’ve loaded the 2010 monthly mean files of potential temperature and salinity as the ds Dataset object, let’s examine its contents.
Note: You can get information about objects and their contents by typing the name of the variable and hittingenterin an interactive session of an IDE such as Spyder or by executing the cell of a Jupyter notebook.
[5]:
ds
[5]:
<xarray.Dataset> Size: 510MB
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50,
k_p1: 51, tile: 13, time: 12, nv: 2, nb: 4)
Coordinates: (12/22)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 200B 0 1 2 3 4 5 6 7 8 ... 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
Zl (k_l) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] 192B dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Z_bnds (k, nv) float32 400B dask.array<chunksize=(50, 2), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
THETA (time, k, tile, j, i) float32 253MB dask.array<chunksize=(1, 25, 7, 45, 45), meta=np.ndarray>
SALT (time, k, tile, j, i) float32 253MB dask.array<chunksize=(1, 25, 7, 45, 45), meta=np.ndarray>
Attributes: (12/62)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
time_coverage_duration: P1M
time_coverage_end: 2010-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2010-01-01T00:00:00
title: ECCO Ocean Temperature and Salinity - Mo...
uuid: f4291248-4181-11eb-82cd-0cc47a3f446dExamining the Dataset object contents¶
Let’s go through ds piece by piece, starting from the top.
1. Object type¶
<xarray.Dataset>
The top line tells us what type of object the variable is. ds is an instance of aDataset defined in xarray.
2. Dimensions¶
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50, k_p1: 51, tile: 13, time: 12, nv: 2, nb: 4)
The Dimensions list shows all of the different dimensions used by all of the different arrays stored in the NetCDF file (and now loaded in the Dataset object).
Arrays may use any combination of these dimensions. In the case of this grid datasets, we find 1D (e.g., depth), 2D (e.g., lat/lon), and 3D (e.g., mask) arrays.
The lengths of these dimensions are next to their name. There are 50 vertical levels in the ECCO v4 model grid, and k corresponds to the vertical dimension centered in the middle of each grid cell. k_u, k_l, and k_p1 are also vertical dimensions, just centered on the bottom, top, and outside of each grid cell respectively. (k_p1 has length 1 greater than the others since it includes both the bottom and top of each grid cell.) i and j correspond to horizontal
dimensions centered in the middle of each grid cell, while i_g and j_g are centered on the u and v faces of each grid cell respectively. The lat-lon-cap grid has 13 tiles. This dataset has 12 monthly-mean records for 2010. The dimension nv is a time dimension that corresponds to the start and end times of the monthly-mean averaging periods. In other words, for every 1 month, there are 2 (nv = 2) time bounds time_bnds, one describing when the month started and the other
when the month ended. SImilarly, the dimension nb corresponds to the horizontal corners of each grid cell, with each grid cell having 4 corners with coordinates given in XC_bnds and YC_bnds.
Note: Each tile in the llc90 grid used by ECCO v4 has 90x90 horizontal grid points. That’s where the 90 in llc90 comes from!
3. Coordinates¶
Some coordinates have an asterisk “*” in front of their names. They are known as dimension coordinates and are always one-dimensional arrays of length 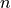 which specify the length of arrays in the dataset in different dimensions.
which specify the length of arrays in the dataset in different dimensions.
Coordinates:
* j (j) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int32 0 1 2 3 4 5 6 7 8 9 ... 80 81 82 83 84 85 86 87 88 89
* k (k) int32 0 1 2 3 4 5 6 7 8 9 ... 40 41 42 43 44 45 46 47 48 49
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2010-12-16T12:00:00
These coordinates are arrays whose values label each grid cell in the arrays. They are used for label-based indexing and alignment.
Let’s look at the three primary spatial coordiates, i, j, k.
[6]:
print(ds.i.long_name)
print(ds.j.long_name)
print(ds.k.long_name)
grid index in x for variables at tracer and 'v' locations
grid index in y for variables at tracer and 'u' locations
grid index in z for tracer variables
i indexes (or labels) the tracer grid cells in the x direction, j indexes the tracer grid cells in the y direction, and similarly k indexes the tracer grid cells in the z direction.
4. Data Variables¶
Data variables:
THETA (time, tile, k, j, i) float32 ...
The Data Variables are one or more xarray.DataArray objects. DataArray objects are labeled, multi-dimensional arrays that may also contain metadata (attributes). DataArray objects are very important to understand because they are container objects which store the numerical arrays of the state estimate fields. We’ll investigate these objects in more detail after completing our survey of this Dataset.
In this NetCDF file one Data variables, THETA, which is stored as a five dimensional array (time, tile, k,j,i) field of average potential temperature. The llc grid has 13 tiles. Each tile has two horizontal dimensions (i,j) and one vertical dimension (k).
THETA is stored here as a 32 bit floating point precision.
Note: The meaning of all MITgcm grid parameters can be found here.
5. Attributes¶
Attributes:
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
creator_email: ecco-group@mit.edu
creator_institution: NASA Jet Propulsion Laboratory (JPL)
creator_name: ECCO Consortium
creator_type: group
creator_url: https://ecco-group.org
The attrs variable is a Python dictionary object containing metadata or any auxilliary information.
Metadata is presented as a set of dictionary key-value pairs. Here the keys are description, A, B, … missing_value. while the values are the corresponding text and non-text values.
To see the metadata value associated with the metadata key called “Conventions” we can print the value as follows:
[7]:
print (ds.attrs['Conventions'])
CF-1.8, ACDD-1.3
“CF-1.8” tells us that ECCO NetCDF output conforms to the Climate and Forecast Conventions version 1.8. How convenient.
Map of the Dataset object¶
Now that we’ve completed our survey, we see that a Dataset is a really a kind of container comprised of (actually pointing to) many other objects.
dims: A
dictthat maps dimension names (keys) with dimension lengths (values)coords: A
dictthat maps dimension names (keys such as k, j, i) with arrays that label each point in the dimension (values)One or more Data Variables that are pointers to
DataArrayobjectsattrs A
dictthat maps different attribute names (keys) with the attributes themselves (values).
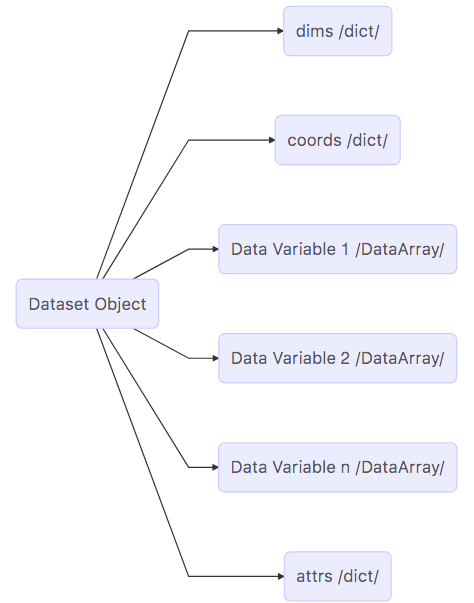
The DataArray Object¶
It is worth looking at the DataArray object in more detail because DataArrays store the arrays that store the ECCO output. Please see the xarray documentation on the DataArray object for more information.
DataArrays are actually very similar to Datasets. They also contain dimensions, coordinates, and attributes. The two main differences between Datasets and DataArrays is that DataArrays have a name (a string) and an array of values. The values array is a numpy n-dimensional array, an ndarray.
Examining the contents of a DataArray¶
Let’s examine the contents of one of the coordinates DataArrays found in ds, XC.
[8]:
ds.XC
[8]:
<xarray.DataArray 'XC' (tile: 13, j: 90, i: 90)> Size: 421kB
dask.array<open_dataset-XC, shape=(13, 90, 90), dtype=float32, chunksize=(13, 90, 90), chunktype=numpy.ndarray>
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
XC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
Attributes:
long_name: longitude of tracer grid cell center
units: degrees_east
coordinate: YC XC
bounds: XC_bnds
comment: nonuniform grid spacing
coverage_content_type: coordinate
standard_name: longitudeExamining the DataArray¶
The layout of DataArrays is very similar to those of Datasets. Let’s examine each part of ds.XC, starting from the top.
1. Object type¶
<xarray.DataArray>
This is indeed a DataArray object from the xarray package.
Note: You can also find the type of an object with the
typecommand:print type(ds.XC)
[9]:
print (type(ds.XC))
<class 'xarray.core.dataarray.DataArray'>
2. Object Name¶
XC
The top line shows DataArray name, XC.
3. Dimensions¶
(tile: 13, j: 90, i: 90)
Unlike THETA, XC does not have time or depth dimensions which makes sense since the longitude of the grid cell centers do not vary with time or depth.
4. The numpy Array¶
array([[[-111.60647 , -111.303 , -110.94285 , ..., 64.791115,
64.80521 , 64.81917 ],
[-104.8196 , -103.928444, -102.87706 , ..., 64.36745 ,
64.41012 , 64.4524 ],
[ -98.198784, -96.788055, -95.14185 , ..., 63.936497,
64.008224, 64.0793 ],
...,
In Dataset objects there are Data variables. In DataArray objects we find numpy arrays. Python prints out a subset of the entire array.
Note:
DataArraysstore only one array whileDataSetscan store one or moreDataArrays.
We access the numpy array by invoking the .values command on the DataArray.
[10]:
print(type(ds.XC.values))
<class 'numpy.ndarray'>
The array that is returned is a numpy n-dimensional array:
[11]:
type(ds.XC.values)
[11]:
numpy.ndarray
Being a numpy array, one can use all of the numerical operations provided by the numpy module on it.
** Note: ** You may find it useful to learn about the operations that can be made on numpy arrays. Here is a quickstart guide: https://docs.scipy.org/doc/numpy-dev/user/quickstart.html
We’ll learn more about how to access the values of this array in a later tutorial. For now it is sufficient to know how to access the arrays!
4. Coordinates¶
The dimensional coordinates (with the asterixes) are
Coordinates:
* j (j) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* i (i) int32 0 1 2 3 4 5 6 7 8 9 10 ... 80 81 82 83 84 85 86 87 88 89
* tile (tile) int32 0 1 2 3 4 5 6 7 8 9 10 11 12
We find three 1D arrays with coordinate labels for j, i, and tile.
[12]:
ds.XC.coords
[12]:
Coordinates:
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
XC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
two other important coordinates here are tile and time
[13]:
print('tile: ')
print(ds.tile.values)
print('time: ')
print(ds.time.values)
tile:
[ 0 1 2 3 4 5 6 7 8 9 10 11 12]
time:
['2010-01-16T12:00:00.000000000' '2010-02-15T00:00:00.000000000'
'2010-03-16T12:00:00.000000000' '2010-04-16T00:00:00.000000000'
'2010-05-16T12:00:00.000000000' '2010-06-16T00:00:00.000000000'
'2010-07-16T12:00:00.000000000' '2010-08-16T12:00:00.000000000'
'2010-09-16T00:00:00.000000000' '2010-10-16T12:00:00.000000000'
'2010-11-16T00:00:00.000000000' '2010-12-16T12:00:00.000000000']
The files we loaded contain the monthly-mean potential temperature and salinity fields for each month in 2010. Here the time coordinates are the center of the averaging periods.
5. Attributes¶
Attributes:
units: degrees_east
long_name: longitude at center of tracer cell
standard_name: longitude_at_c_location
valid_range: -180., 180.
The XC variable has a long_name (longitude at center of tracer cell) and units (degrees_east) and other information. This metadata was loaded from the NetCDF file. The entire attribute dictionary is accessed using .attrs.
[14]:
ds.XC.attrs
[14]:
{'long_name': 'longitude of tracer grid cell center',
'units': 'degrees_east',
'coordinate': 'YC XC',
'bounds': 'XC_bnds',
'comment': 'nonuniform grid spacing',
'coverage_content_type': 'coordinate',
'standard_name': 'longitude'}
[15]:
ds.XC.attrs['units']
[15]:
'degrees_east'
Map of the DataArray Object¶
The DataArray can be mapped out with the following diagram:
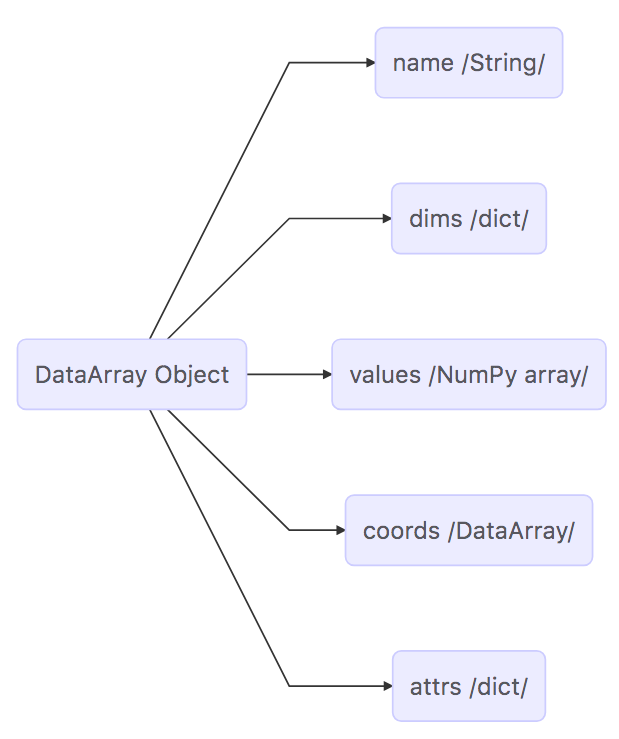
Summary¶
Now you know the basics of the Dataset and DataArray objects that will store the ECCO v4 model grid parameters and state estimate output variables. Go back and take a look athe grid  object that we originally loaded. It should make a lot more sense now!
object that we originally loaded. It should make a lot more sense now!